The randomized SVD and its relatives are workhorse algorithms for low-rank approximation. In this post, I hope to provide a fairly brief introduction to this useful method. In the following post, we’ll look into its analysis.
The randomized SVD is dead-simple to state: To approximate an ![]() matrix
matrix ![]() by a rank-
by a rank-![]() matrix, perform the following steps:
matrix, perform the following steps:
- Collect information. Form
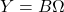 where
where 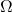 is an appropriately chosen
is an appropriately chosen 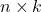 random matrix.
random matrix. - Orthogonalize. Compute an orthonormal basis
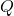 for the column space of
for the column space of 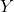 by, e.g., thin QR factorization.
by, e.g., thin QR factorization. - Project. Form
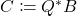 . (Here,
. (Here,  denotes the conjugate transpose of a complex matrix, which reduces to the ordinary transpose if the matrix is real.)
denotes the conjugate transpose of a complex matrix, which reduces to the ordinary transpose if the matrix is real.)
If all one cares about is a low-rank approximation to the matrix ![]() , one can stop the randomized SVD here, having obtained the approximation
, one can stop the randomized SVD here, having obtained the approximation
![]()
As the name “randomized SVD” suggests, one can easily “upgrade” the approximation ![]() to a compact SVD format:
to a compact SVD format:
- SVD. Compute a compact SVD of the matrix
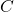 :
: 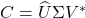 where
where 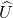 and
and 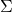 and
and 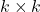 and
and 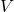 is
is 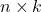 .
. - Clean up. Set
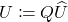 .
.
We now have approximated ![]() by a rank-
by a rank-![]() matrix
matrix ![]() in compact SVD form:
in compact SVD form:
![]()
One can use the factors ![]() ,
, ![]() , and
, and ![]() of the approximation
of the approximation ![]() as estimates of the singular vectors and values of the matrix
as estimates of the singular vectors and values of the matrix ![]() .
.
What Can You Do with the Randomized SVD?
The randomized SVD has many uses. Pretty much anywhere you would ordinarily use an SVD is a candidate for the randomized SVD. Applications include model reduction, fast direct solvers, computational biology, uncertainty quantification, among numerous others.
To speak in broad generalities, there are two ways to use the randomized SVD:
- As an approximation. First, we could use
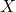 as a compressed approximation to
as a compressed approximation to 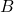 . Using the format
. Using the format 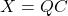 ,
, 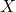 requires just
requires just 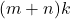 numbers of storage, whereas
numbers of storage, whereas 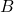 requires a much larger
requires a much larger 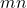 numbers of storage. As I detailed at length in my blog post on low-rank matrices, many operations are cheap for the low-rank matrix
numbers of storage. As I detailed at length in my blog post on low-rank matrices, many operations are cheap for the low-rank matrix 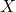 . For instance, we can compute a matrix–vector product
. For instance, we can compute a matrix–vector product 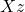 in roughly
in roughly 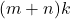 operations rather than the
operations rather than the 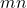 operations to compute
operations to compute 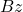 . For these use cases, we don’t need the “SVD” part of the randomized SVD.
. For these use cases, we don’t need the “SVD” part of the randomized SVD. - As an approximate SVD. Second, we can actually use the
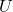 ,
, 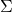 , and
, and 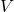 produced by the randomized SVD as approximations to the singular vectors and values of
produced by the randomized SVD as approximations to the singular vectors and values of 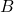 . In these applications, we should be careful. Just because
. In these applications, we should be careful. Just because 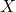 is a good approximation to
is a good approximation to 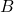 , it is not necessarily the case that
, it is not necessarily the case that 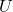 ,
, 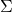 , and
, and 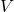 are close to the singular vectors and values of
are close to the singular vectors and values of 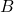 . To use the randomized SVD in this context, it is safest to use posterior diagnostics (such as the ones developed in this paper of mine) to ensure that the singular values/vectors of interest are computed to a high enough accuracy. A useful rule of thumb is the smallest two to five singular values and vectors computed by the randomized SVD are suspect and should be used in applications only with extreme caution. When the appropriate care is taken and for certain problems, the randomized SVD can provide accurate singular vectors far faster than direct methods.
. To use the randomized SVD in this context, it is safest to use posterior diagnostics (such as the ones developed in this paper of mine) to ensure that the singular values/vectors of interest are computed to a high enough accuracy. A useful rule of thumb is the smallest two to five singular values and vectors computed by the randomized SVD are suspect and should be used in applications only with extreme caution. When the appropriate care is taken and for certain problems, the randomized SVD can provide accurate singular vectors far faster than direct methods.
Accuracy of the Randomized SVD
How accurate is the approximation ![]() produced by the randomized SVD? No rank-
produced by the randomized SVD? No rank-![]() approximation can be more accurate than the best rank-
approximation can be more accurate than the best rank-![]() approximation
approximation ![]() to the matrix
to the matrix ![]() , furnished by an exact SVD of
, furnished by an exact SVD of ![]() truncated to rank
truncated to rank ![]() . Thus, we’re interested in how much larger
. Thus, we’re interested in how much larger ![]() is than
is than ![]() .
.
Frobenius Norm Error
A representative result (see, e.g., Theorem 8.1 of this paper) describing the error of the randomized SVD is the following:
(1) ![]()
This result states that the average squared Frobenius-norm error of the randomized SVD is comparable with the best rank-![]() approximation error for any
approximation error for any ![]() . In particular, choosing
. In particular, choosing ![]() , we see that the randomized SVD error is at most
, we see that the randomized SVD error is at most ![]() times the best rank-
times the best rank-![]() approximation
approximation
(2) ![]()
Choosing ![]() , we see that the randomized SVD has at most twice the error of the best rank-
, we see that the randomized SVD has at most twice the error of the best rank-![]() approximation
approximation
(3) ![]()
In effect, these results tell us that if we want an approximation that is nearly as good as the best rank-![]() approximation, using an approximation rank of, say,
approximation, using an approximation rank of, say, ![]() or
or ![]() for the randomized SVD suffices. These results hold even for worst-case matrices. For nice matrices with steadily decaying singular values, the randomized SVD can perform even better than equations (2)–(3) would suggest.
for the randomized SVD suffices. These results hold even for worst-case matrices. For nice matrices with steadily decaying singular values, the randomized SVD can perform even better than equations (2)–(3) would suggest.
Spectral Norm Error
The spectral norm is often a better error metric for matrix computations than the Frobenius norm. Is the randomized SVD also comparable with the best rank-![]() approximation when we use the spectral norm? In this setting, a representative error bound (see, e.g., Theorem 8.7 of this paper) is
approximation when we use the spectral norm? In this setting, a representative error bound (see, e.g., Theorem 8.7 of this paper) is
(4) ![]()
The spectral norm error of the randomized SVD depends on the Frobenius norm error of the best rank-![]() approximation for
approximation for ![]() .
.
Recall that the spectral and Frobenius norm can be defined in terms of the singular values of the matrix ![]() :
:
![]()
Rewriting (4) using these formulas, we get
![Rendered by QuickLaTeX.com \[\mathbb{E} \left\|B - X\right\|^2 \le \min_{r \le k-2} \left( 1 + \frac{2r}{k-(r+1)} \right) \left(\sigma_{r+1}^2 + \frac{\mathrm{e}^2}{k-r} \sum_{i>r} \sigma_i^2 \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-516cee5dcd03a69b94d82e30744a0782_l3.png)
Despite being interested in the largest singular value of the error matrix ![]() , this bound demonstrates that the randomized SVD incurs errors based on the entire tail of
, this bound demonstrates that the randomized SVD incurs errors based on the entire tail of ![]() ‘s singular values
‘s singular values ![]() . The randomized SVD is much worse than the best rank-
. The randomized SVD is much worse than the best rank-![]() approximation for a matrix
approximation for a matrix ![]() with a long detail of slowly declining singular values.
with a long detail of slowly declining singular values.
Improving the Approximation: Subspace Iteration and Block Krylov Iteration
We saw that the randomized SVD produces nearly optimal low-rank approximations when we measure using the Frobenius norm. When we measure using the spectral norm, we have a split decision: If the singular values decay rapidly, the randomized SVD is near-optimal; if the singular values decay more slowly, the randomized SVD can suffer from large errors.
Fortunately, there are two fixes to improve the randomized SVD in the presence of slowly decaying singular values:
- Subspace iteration: To improve the quality of the randomized SVD, we can combine it with the famous power method for eigenvalue computations. Instead of
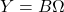 , we use the powered expression
, we use the powered expression 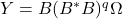 . Powering has the effect of amplifying the large singular values of
. Powering has the effect of amplifying the large singular values of 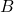 and dimishing the influence of the tail.
and dimishing the influence of the tail. - Block Krylov iteration: Subspace iteration is effective, but fairly wasteful. To compute
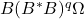 we compute the block Krylov sequence
we compute the block Krylov sequence
and throw away all but the last term. To make more economical use of resources, we can use the entire sequence as our information matrix![Rendered by QuickLaTeX.com \[B\Omega, BB^*B\Omega, BB^*BB^*B\Omega,\ldots,B(B^*B)^q\Omega\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-e8f795167565126cb4a6f1732351b1ef_l3.png)
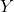 :
:
Storing the entire block Krylov sequence is more memory-intensive but makes more efficient use of matrix products than subspace iteration.![Rendered by QuickLaTeX.com \[Y = \begin{bmatrix} B\Omega & BB^*B\Omega & BB^*BB^*B\Omega& \cdots & B(B^*B)^q\Omega\end{bmatrix}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-2ab5359def6111f87fdf8081ed740642_l3.png)
Both subspace iteration and block Krylov iteration should be carefully implemented to produce accurate results.
Both subspace iteration and block Krylov iteration diminish the effect of a slowly decaying tail of singular values on the accuracy of the randomized SVD. The more slowly the tail decays, the larger one must take the number of iterations ![]() to obtain accurate results.
to obtain accurate results.
Dear Mr. Epperly,
Could you please provide references for the formulas mentioned in this post?
Kind regards,
Eva-Maria Haslhofer
I can’t believe I didn’t have references already! Results of the form given in this post go back to the famous paper of Halko, Martinsson, and Tropp (2011). The precise versions given here appear as Theorems 8.1 and 8.7 in Webber & Tropp (2023).
Thank you very much for the references and for the excellent introduction to the topic!