Consider two complex-valued square matrices ![]() and
and ![]() . The first matrix
. The first matrix ![]() is Hermitian, being equal
is Hermitian, being equal ![]() to its conjugate transpose
to its conjugate transpose ![]() . The other matrix
. The other matrix ![]() is non-Hermitian,
is non-Hermitian, ![]() . Let’s see how long it takes to compute their eigenvalue decompositions in MATLAB:
. Let’s see how long it takes to compute their eigenvalue decompositions in MATLAB:
>> A = randn(1e3) + 1i*randn(1e3); A = (A+A')/2;
>> tic; [V_A,D_A] = eig(A); toc % Hermitian matrix
Elapsed time is 0.415145 seconds.
>> B = randn(1e3) + 1i*randn(1e3);
>> tic; [V_B,D_B] = eig(B); toc % non-Hermitian matrix
Elapsed time is 1.668246 seconds.We see that it takes ![]() longer to compute the eigenvalues of the non-Hermitian matrix
longer to compute the eigenvalues of the non-Hermitian matrix ![]() as compared to the Hermitian matrix
as compared to the Hermitian matrix ![]() . Moreover, the matrix
. Moreover, the matrix ![]() of eigenvectors for a Hermitian matrix
of eigenvectors for a Hermitian matrix ![]() is a unitary matrix,
is a unitary matrix, ![]() .
.
There are another class of matrices with nice eigenvalue decompositions, normal matrices. A square, complex-valued matrix ![]() is normal if
is normal if ![]() . The matrix
. The matrix ![]() of eigenvectors for a normal matrix
of eigenvectors for a normal matrix ![]() is also unitary,
is also unitary, ![]() . An important class of normal matrices are unitary matrices themselves. A unitary matrix
. An important class of normal matrices are unitary matrices themselves. A unitary matrix ![]() is always normal since it satisfies
is always normal since it satisfies ![]() .
.
Let’s see how long it takes MATLAB to compute the eigenvalue decomposition of a unitary (and thus normal) matrix:
>> U = V_A; % unitary, and thus normal, matrix
>> tic; [V_U,D_U] = eig(U); toc % normal matrix
Elapsed time is 2.201017 seconds.Even longer than it took to compute an eigenvalue decomposition of the non-normal matrix ![]() ! Can we make the normal eigenvalue decomposition closer to the speed of the Hermitian eigenvalue decomposition?
! Can we make the normal eigenvalue decomposition closer to the speed of the Hermitian eigenvalue decomposition?
Here is the start of an idea. Every square matrix ![]() has a Cartesian decomposition:
has a Cartesian decomposition:
![]()
For a normal matrix ![]() , the Hermitian and skew-Hermitian parts commute,
, the Hermitian and skew-Hermitian parts commute, ![]() . We know from matrix theory that commuting Hermitian matrices are simultaneously diagonalizable, i.e., there exists
. We know from matrix theory that commuting Hermitian matrices are simultaneously diagonalizable, i.e., there exists ![]() such that
such that ![]() and
and ![]() for diagonal matrices
for diagonal matrices ![]() and
and ![]() . Thus, given access to such
. Thus, given access to such ![]() ,
, ![]() has eigenvalue decomposition
has eigenvalue decomposition
![]()
Here’s a first attempt to turn this insight into an algorithm. First, compute the Hermitian part ![]() of
of ![]() , diagonalize
, diagonalize ![]() , and then see if
, and then see if ![]() diagonalizes
diagonalizes ![]() . Let’s test this out on a
. Let’s test this out on a ![]() example:
example:
>> C = orth(randn(2) + 1i*randn(2)); % unitary matrix
>> H = (C+C')/2; % Hermitian part
>> [Q,~] = eig(H);
>> Q'*C*Q % check to see if diagonal
ans =
-0.9933 + 0.1152i -0.0000 + 0.0000i
0.0000 + 0.0000i -0.3175 - 0.9483iYay! We’ve succeeded at diagonalizing the matrix ![]() using only a Hermitian eigenvalue decomposition. But we should be careful about declaring victory too early. Here’s a bad example:
using only a Hermitian eigenvalue decomposition. But we should be careful about declaring victory too early. Here’s a bad example:
>> C = [1 1i;1i 1]; % normal matrix
>> H = (C+C')/2;
>> [Q,~] = eig(H);
>> Q'*C*Q % oh no! not diagonal
ans =
1.0000 + 0.0000i 0.0000 + 1.0000i
0.0000 + 1.0000i 1.0000 + 0.0000iWhat’s going on here? The issue is that the Hermitian part ![]() for this matrix has a repeated eigenvalue. Thus,
for this matrix has a repeated eigenvalue. Thus, ![]() has multiple different valid matrices of eigenvectors. (In this specific case, every unitary matrix
has multiple different valid matrices of eigenvectors. (In this specific case, every unitary matrix ![]() diagonalizes
diagonalizes ![]() .) By looking at
.) By looking at ![]() alone, we don’t know which
alone, we don’t know which ![]() matrix to pick which also diagonalizes
matrix to pick which also diagonalizes ![]() .
.
He and Kressner developed a beautifully simple randomized algorithm called RandDiag to circumvent this failure mode. The idea is straightforward:
- Form a random linear combination
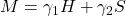 of the Hermitian and skew-Hermitian parts of
of the Hermitian and skew-Hermitian parts of  , with standard normal random coefficients
, with standard normal random coefficients 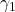 and
and 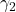 .
. - Compute
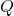 that diagonalizes
that diagonalizes 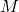 .
.
That’s it!
To get a sense of why He and Kressner’s algorithm works, suppose that ![]() has some repeated eigenvalues and
has some repeated eigenvalues and ![]() has all distinct eigenvalues. Given this setup, it seems likely that a random linear combination of
has all distinct eigenvalues. Given this setup, it seems likely that a random linear combination of ![]() and
and ![]() will also have all distinct eigenvalues. (It would take a very special circumstances for a random linear combination to yield two eigenvalues that are exactly the same!) Indeed, this intuition is a fact: With 100% probability,
will also have all distinct eigenvalues. (It would take a very special circumstances for a random linear combination to yield two eigenvalues that are exactly the same!) Indeed, this intuition is a fact: With 100% probability, ![]() diagonalizing a Gaussian random linear combination of simultaneously diagonalizable matrices
diagonalizing a Gaussian random linear combination of simultaneously diagonalizable matrices ![]() and
and ![]() also diagonalizes
also diagonalizes ![]() and
and ![]() individually.
individually.
MATLAB code for RandDiag is as follows:
function Q = rand_diag(C)
H = (C+C')/2; S = (C-C')/2i;
M = randn*H + randn*S;
[Q,~] = eig(M);
endWhen applied to our hard ![]() example from before, RandDiag succeeds at giving a matrix that diagonalizes
example from before, RandDiag succeeds at giving a matrix that diagonalizes ![]() :
:
>> Q = rand_diag(C);
>> Q'*C*Q
ans =
1.0000 - 1.0000i -0.0000 + 0.0000i
-0.0000 - 0.0000i 1.0000 + 1.0000iFor computing the matrix of eigenvectors for a ![]() unitary matrix, RandDiag takes 0.4 seconds, just as fast as the Hermitian eigendecomposition did.
unitary matrix, RandDiag takes 0.4 seconds, just as fast as the Hermitian eigendecomposition did.
>> tic; V_U = rand_diag(U); toc
Elapsed time is 0.437309 seconds.He and Kressner’s algorithm is delightful. Ultimately, it uses randomness in only a small way. For most coefficients ![]() , a matrix
, a matrix ![]() diagonalizing
diagonalizing ![]() will also diagonalize
will also diagonalize ![]() . But, for any specific choice of
. But, for any specific choice of ![]() , there is a possibility of failure. To avoid this possibility, we can just pick
, there is a possibility of failure. To avoid this possibility, we can just pick ![]() and
and ![]() at random. It’s really as simple as that.
at random. It’s really as simple as that.
References: RandDiag was proposed in A simple, randomized algorithm for diagonalizing normal matrices by He and Kressner (2024), building on their earlier work in Randomized Joint Diagonalization of Symmetric Matrices (2022) which considers the general case of using random linear combinations to (approximately) simultaneous diagonalize (nearly) commuting matrices. RandDiag is an example of a linear algebraic algorithm that uses randomness to put the input into “general position”; see Randomized matrix computations: Themes and variations by Kireeva and Tropp (2024) for a discussion of this, and other, ways of using randomness to design matrix algorithms.