I’m excited to share that my paper, Fast and forward stable randomized algorithms for linear least-squares problems has been released as a preprint on arXiv.
With the release of this paper, now seemed like a great time to discuss a topic I’ve been wanting to write about for a while: sketching. For the past two decades, sketching has become a widely used algorithmic tool in matrix computations. Despite this long history, questions still seem to be lingering about whether sketching really works:
In this post, I want to take a critical look at the question “does sketching work”? Answering this question requires answering two basic questions:
- What is sketching?
- What would it mean for sketching to work?
I think a large part of the disagreement over the efficacy of sketching boils down to different answers to these questions. By considering different possible answers to these questions, I hope to provide a balanced perspective on the utility of sketching as an algorithmic primitive for solving linear algebra problems.
Sketching
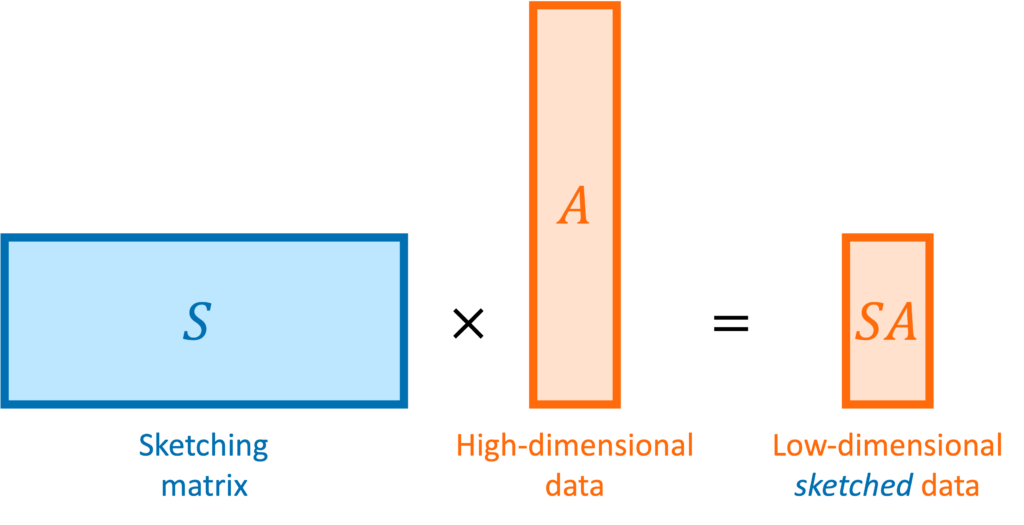
In matrix computations, sketching is really a synonym for (linear) dimensionality reduction. Suppose we are solving a problem involving one or more high-dimensional vectors 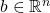 or perhaps a tall matrix
or perhaps a tall matrix 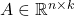 . A sketching matrix is a
. A sketching matrix is a 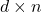 matrix
matrix 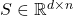 where
where 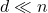 . When multiplied into a high-dimensional vector
. When multiplied into a high-dimensional vector 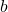 or tall matrix
or tall matrix  , the sketching matrix
, the sketching matrix 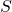 produces compressed or “sketched” versions
produces compressed or “sketched” versions 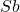 and
and 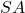 that are much smaller than the original vector
that are much smaller than the original vector 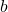 and matrix
and matrix  .
.
Let 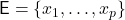 be a collection of vectors. For
be a collection of vectors. For 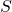 to be a “good” sketching matrix for
to be a “good” sketching matrix for 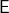 , we require that
, we require that 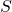 preserves the lengths of every vector in
preserves the lengths of every vector in 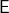 up to a distortion parameter
up to a distortion parameter 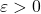 :
:
(1) ![Rendered by QuickLaTeX.com \[(1-\varepsilon) \norm{x}\le\norm{Sx}\le(1+\varepsilon)\norm{x} \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-735df49a45ff5d0bb27d478569004bdd_l3.png)
for every
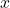
in
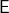
.
For linear algebra problems, we often want to sketch a matrix  . In this case, the appropriate set
. In this case, the appropriate set 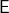 that we want our sketch to be “good” for is the column space of the matrix
that we want our sketch to be “good” for is the column space of the matrix  , defined to be
, defined to be
![Rendered by QuickLaTeX.com \[\operatorname{col}(A) \coloneqq \{ Ax : x \in \real^k \}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-5ecde6b43df29caba14fd43682b4cc86_l3.png)
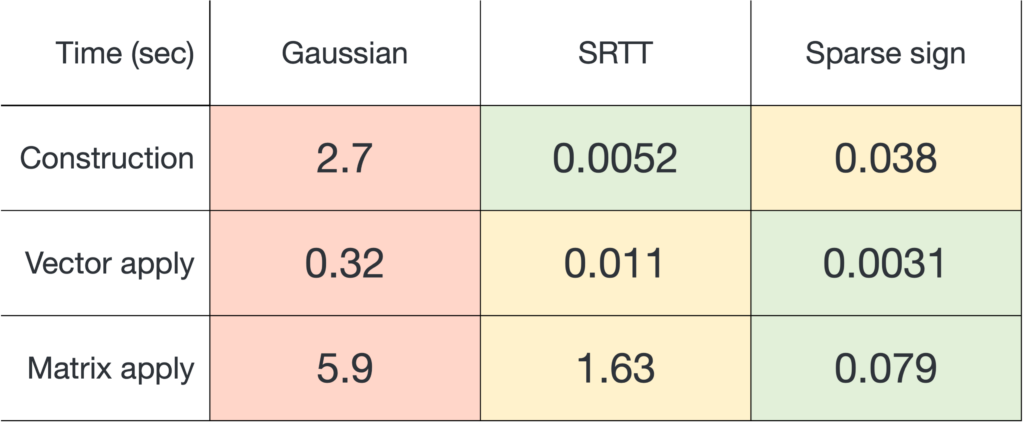
Remarkably, there exist many sketching matrices that achieve distortion

for
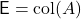
with an output dimension of roughly
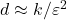
. In particular, the sketching dimension
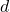
is proportional to the number of columns
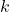
of

. This is pretty neat! We can design a single sketching matrix
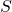
which preserves the lengths of all infinitely-many vectors
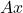
in the column space of

.
Sketching Matrices
There are many types of sketching matrices, each with different benefits and drawbacks. Many sketching matrices are based on randomized constructions in the sense that entries of 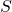 are chosen to be random numbers. Broadly, sketching matrices can be classified into two types:
are chosen to be random numbers. Broadly, sketching matrices can be classified into two types:
- Data-dependent sketches. The sketching matrix
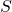 is constructed to work for a specific set of input vectors
is constructed to work for a specific set of input vectors 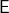 .
. - Oblivious sketches. The sketching matrix
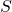 is designed to work for an arbitrary set of input vectors
is designed to work for an arbitrary set of input vectors 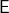 of a given size (i.e.,
of a given size (i.e., 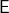 has
has 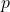 elements) or dimension (
elements) or dimension (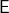 is a
is a 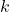 -dimensional linear subspace).
-dimensional linear subspace).
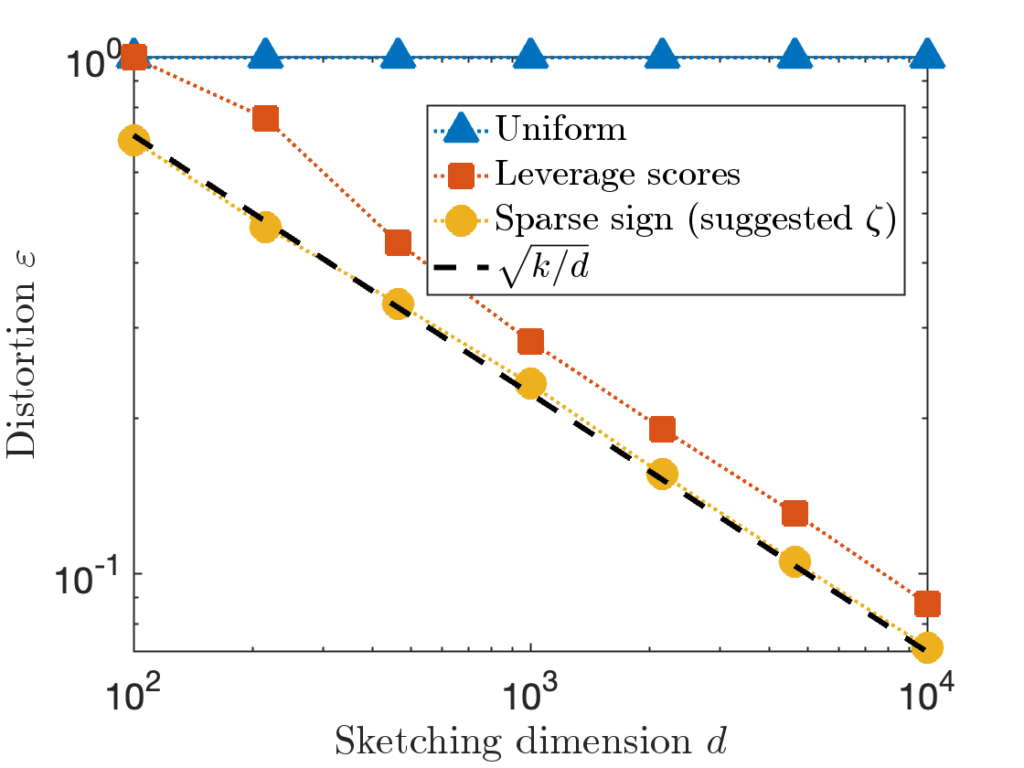
We will only discuss oblivious sketching for this post. We will look at three types of sketching matrices: Gaussian embeddings, subsampled randomized trignometric transforms, and sparse sign embeddings.
The details of how these sketching matrices are built and their strengths and weaknesses can be a little bit technical. All three constructions are independent from the rest of this article and can be skipped on a first reading. The main point is that good sketching matrices exist and are fast to apply: Reducing 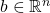 to
to 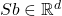 requires roughly
requires roughly 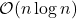 operations, rather than the
operations, rather than the 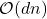 operations we would expect to multiply a
operations we would expect to multiply a 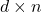 matrix and a vector of length
matrix and a vector of length 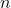 .
.
The simplest type of sketching matrix 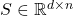 is obtained by (independently) setting every entry of
is obtained by (independently) setting every entry of 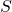 to be a Gaussian random number with mean zero and variance
to be a Gaussian random number with mean zero and variance 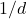 . Such a sketching matrix is called a Gaussian embedding.
. Such a sketching matrix is called a Gaussian embedding.
Benefits. Gaussian embeddings are simple to code up, requiring only a standard matrix product to apply to a vector 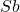 or matrix
or matrix 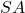 . Gaussian embeddings admit a clean theoretical analysis, and their mathematical properties are well-understood.
. Gaussian embeddings admit a clean theoretical analysis, and their mathematical properties are well-understood.
Drawbacks. Computing 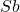 for a Gaussian embedding costs
for a Gaussian embedding costs 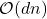 operations, significantly slower than the other sketching matrices we will consider below. Additionally, generating and storing a Gaussian embedding can be computationally expensive.
operations, significantly slower than the other sketching matrices we will consider below. Additionally, generating and storing a Gaussian embedding can be computationally expensive.
The subsampled randomized trigonometric transform (SRTT) sketching matrix takes a more complicated form. The sketching matrix is defined to be a scaled product of three matrices
![Rendered by QuickLaTeX.com \[S = \sqrt{\frac{n}{d}} \cdot R \cdot F \cdot D.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-92095cbf9c7d55273bd572a33debe415_l3.png)
These matrices have the following definitions:
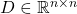 is a diagonal matrix whose entries are each a random
is a diagonal matrix whose entries are each a random 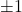 (chosen independently with equal probability).
(chosen independently with equal probability).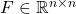 is a fast trigonometric transform such as a fast discrete cosine transform.
is a fast trigonometric transform such as a fast discrete cosine transform.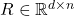 is a selection matrix. To generate
is a selection matrix. To generate 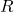 , let
, let  be a random subset of
be a random subset of  , selected without replacement.
, selected without replacement. 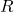 is defined to be a matrix for which
is defined to be a matrix for which 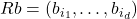 for every vector
for every vector 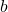 .
.
To store 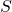 on a computer, it is sufficient to store the
on a computer, it is sufficient to store the 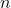 diagonal entries of
diagonal entries of 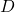 and the
and the 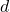 selected coordinates
selected coordinates  defining
defining 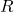 . Multiplication of
. Multiplication of 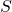 against a vector
against a vector 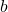 should be carried out by applying each of the matrices
should be carried out by applying each of the matrices 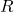 ,
, 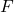 , and
, and 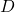 in sequence, such as in the following MATLAB code:
in sequence, such as in the following MATLAB code:
% Generate randomness for S
signs = 2*randi(2,m,1)-3; % diagonal entries of D
idx = randsample(m,d); % indices i_1,...,i_d defining R
% Multiply S against b
c = signs .* b % multiply by D
c = dct(c) % multiply by F
c = c(idx) % multiply by R
c = sqrt(n/d) * c % scale
Benefits. 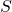 can be applied to a vector
can be applied to a vector 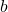 in
in 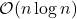 operations, a significant improvement over the
operations, a significant improvement over the 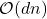 cost of a Gaussian embedding. The SRTT has the lowest memory and random number generation requirements of any of the three sketches we discuss in this post.
cost of a Gaussian embedding. The SRTT has the lowest memory and random number generation requirements of any of the three sketches we discuss in this post.
Drawbacks. Applying 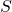 to a vector requires a good implementation of a fast trigonometric transform. Even with a high-quality trig transform, SRTTs can be significantly slower than sparse sign embeddings (defined below). SRTTs are hard to parallelize. In theory, the sketching dimension should be chosen to be
to a vector requires a good implementation of a fast trigonometric transform. Even with a high-quality trig transform, SRTTs can be significantly slower than sparse sign embeddings (defined below). SRTTs are hard to parallelize. In theory, the sketching dimension should be chosen to be 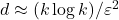 , larger than for a Gaussian sketch.
, larger than for a Gaussian sketch.
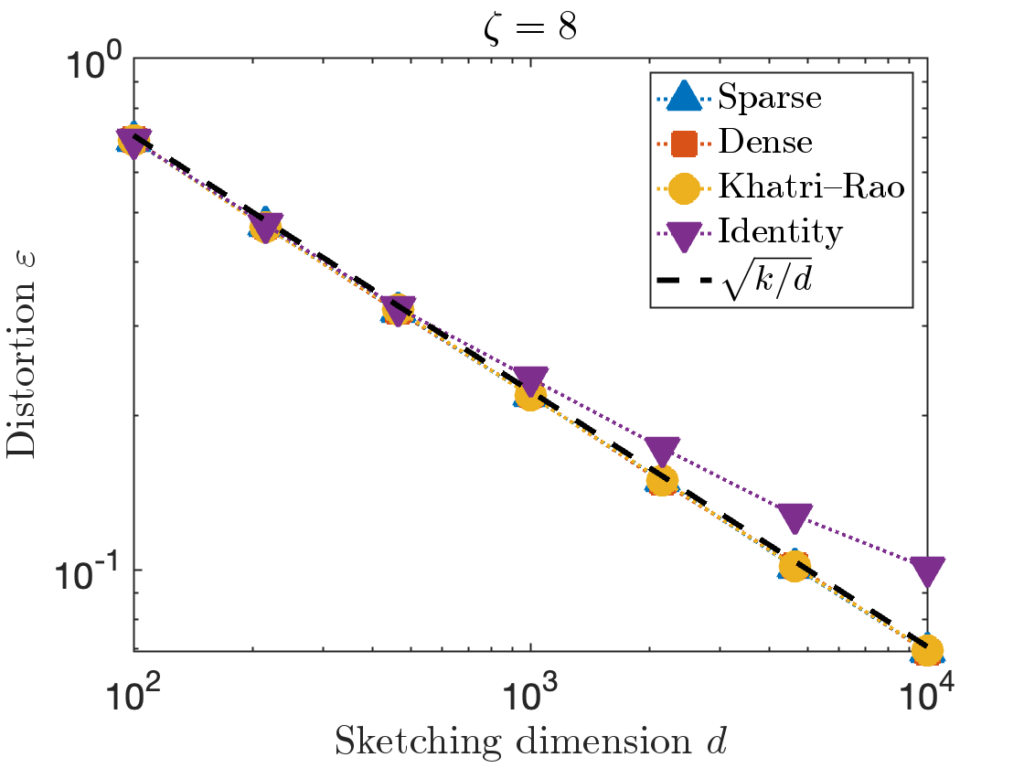
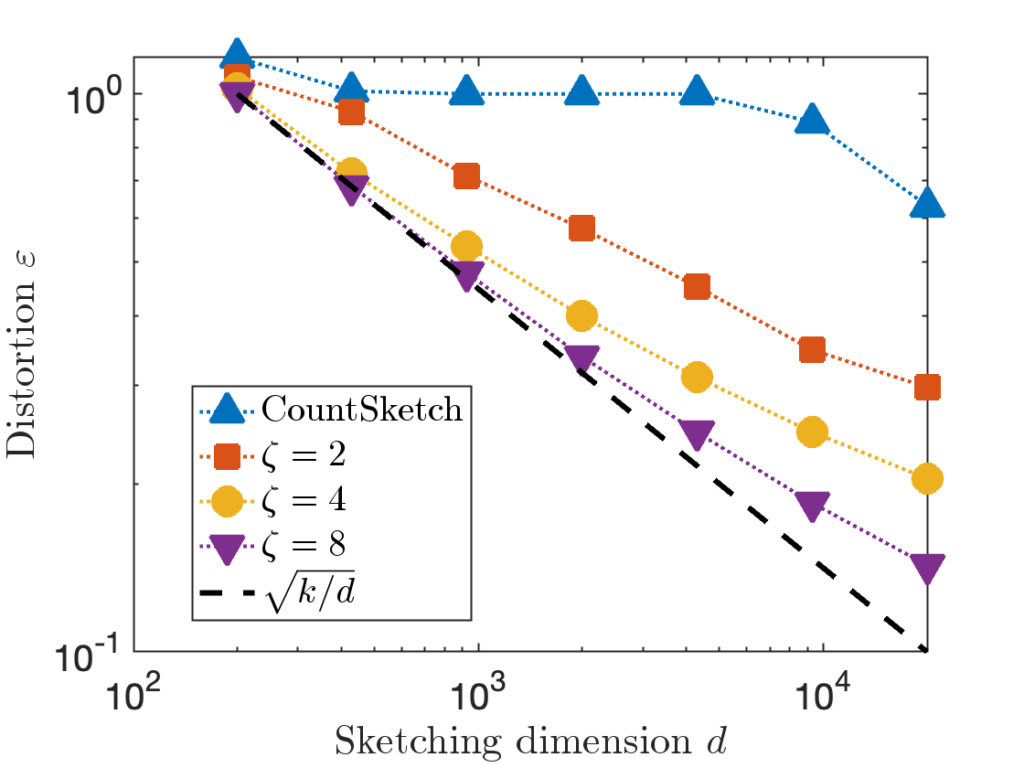
A sparse sign embedding takes the form
![Rendered by QuickLaTeX.com \[S = \frac{1}{\sqrt{\zeta}} \begin{bmatrix} s_1 & s_2 & \cdots & s_n \end{bmatrix}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-bb5806806e2e5900d4351c08befbd82e_l3.png)
Here, each column
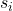
is an independently generated random vector with exactly
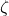
nonzero entries with random
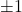
values in uniformly random positions. The result is a
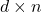
matrix with only
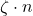
nonzero entries. The parameter
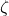
is often set to a small constant like
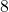
in practice.
Benefits. By using a dedicated sparse matrix library, 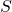 can be very fast to apply to a vector
can be very fast to apply to a vector 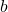 (either
(either 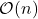 or
or 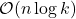 operations) to apply to a vector, depending on parameter choices (see below). With a good sparse matrix library, sparse sign embeddings are often the fastest sketching matrix by a wide margin.
operations) to apply to a vector, depending on parameter choices (see below). With a good sparse matrix library, sparse sign embeddings are often the fastest sketching matrix by a wide margin.
Drawbacks. To be fast, sparse sign embeddings requires a good sparse matrix library. They require generating and storing roughly 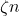 random numbers, higher than SRTTs (roughly
random numbers, higher than SRTTs (roughly 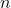 numbers) but much less than Gaussian embeddings (
numbers) but much less than Gaussian embeddings (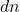 numbers). In theory, the sketching dimension should be chosen to be
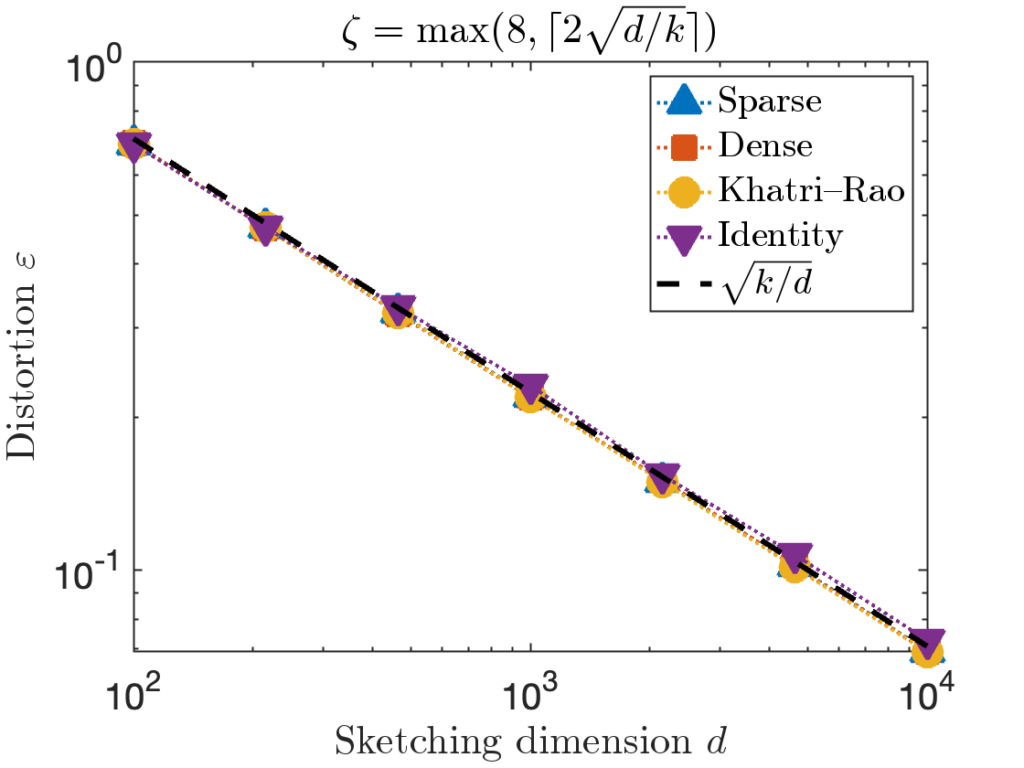
numbers). In theory, the sketching dimension should be chosen to be 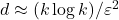 and the sparsity should be set to
and the sparsity should be set to 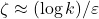 ; the theoretically sanctioned sketching dimension (at least according to existing theory) is larger than for a Gaussian sketch. In practice, we can often get away with using
; the theoretically sanctioned sketching dimension (at least according to existing theory) is larger than for a Gaussian sketch. In practice, we can often get away with using 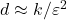 and
and 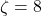 .
.
Summary
Using either SRTTs or sparse maps, a sketching a vector of length 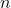 down to
down to 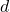 dimensions requires only
dimensions requires only 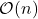 to
to 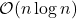 operations. To apply a sketch to an entire
operations. To apply a sketch to an entire 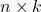 matrix
matrix  thus requires roughly
thus requires roughly 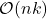 operations. Therefore, sketching offers the promise of speeding up linear algebraic computations involving
operations. Therefore, sketching offers the promise of speeding up linear algebraic computations involving  , which typically take
, which typically take 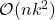 operations.
operations.
How Can You Use Sketching?
The simplest way to use sketching is to first apply the sketch to dimensionality-reduce all of your data and then apply a standard algorithm to solve the problem using the reduced data. This approach to using sketching is called sketch-and-solve.
As an example, let’s apply sketch-and-solve to the least-squares problem:
(2) ![Rendered by QuickLaTeX.com \[\operatorname*{minimize}_{x\in\real^k} \norm{Ax - b}. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-07528304ce875f54aa1173062c5ec10a_l3.png)
We assume this problem is highly overdetermined with

having many more rows
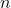
than columns
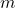
.
To solve this problem with sketch-and-solve, generate a good sketching matrix 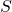 for the set
for the set 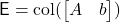 . Applying
. Applying 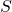 to our data
to our data  and
and 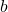 , we get a dimensionality-reduced least-squares problem
, we get a dimensionality-reduced least-squares problem
(3) ![Rendered by QuickLaTeX.com \[\operatorname*{minimize}_{\hat{x}\in\real^k} \norm{(SA)\hat{x} - Sb}. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7110d3e9c1dd7e29793603a5bafdd201_l3.png)
The solution
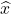
is the sketch-and-solve solution to the least-squares problem, which we can use as an approximate solution to the original least-squares problem.
Least-squares is just one example of the sketch-and-solve paradigm. We can also use sketching to accelerate other algorithms. For instance, we could apply sketch-and-solve to clustering. To cluster data points  , first apply sketching to obtain
, first apply sketching to obtain  and then apply an out-of-the-box clustering algorithms like k-means to the sketched data points.
and then apply an out-of-the-box clustering algorithms like k-means to the sketched data points.
Does Sketching Work?
Most often, when sketching critics say “sketching doesn’t work”, what they mean is “sketch-and-solve doesn’t work”.
To address this question in a more concrete setting, let’s go back to the least-squares problem (2). Let 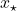 denote the optimal least-squares solution and let
denote the optimal least-squares solution and let 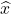 be the sketch-and-solve solution (3). Then, using the distortion condition (1), one can show that
be the sketch-and-solve solution (3). Then, using the distortion condition (1), one can show that
![Rendered by QuickLaTeX.com \[\norm{A\hat{x} - b} \le \frac{1+\varepsilon}{1-\varepsilon} \norm{Ax - b}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-3d1d034bd8bf93f1c15e435a27a355ee_l3.png)
If we use a sketching matrix with a distortion of
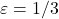
, then this bound tells us that
(4) ![Rendered by QuickLaTeX.com \[\norm{A\hat{x} - b} \le 2\norm{Ax_\star - b}. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-588170743415f491f48ef2adc3d46628_l3.png)
Is this a good result or a bad result? Ultimately, it depends. In some applications, the quality of a putative least-squares solution 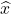 is can be assessed from the residual norm
is can be assessed from the residual norm 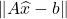 . For such applications, the bound (4) ensures that
. For such applications, the bound (4) ensures that 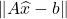 is at most twice
is at most twice 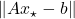 . Often, this means
. Often, this means 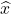 is a pretty decent approximate solution to the least-squares problem.
is a pretty decent approximate solution to the least-squares problem.
For other problems, the appropriate measure of accuracy is the so-called forward error 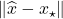 , measuring how close
, measuring how close 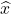 is to
is to 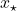 . For these cases, it is possible that
. For these cases, it is possible that 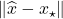 might be large even though the residuals are comparable (4).
might be large even though the residuals are comparable (4).
Let’s see an example, using the MATLAB code from my paper:
[A, b, x, r] = random_ls_problem(1e4, 1e2, 1e8, 1e-4); % Random LS problem
S = sparsesign(4e2, 1e4, 8); % Sparse sign embedding
sketch_and_solve = (S*A) \ (S*b); % Sketch-and-solve
direct = A \ b; % MATLAB mldivide
Here, we generate a random least-squares problem of size 10,000 by 100 (with condition number 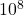 and residual norm
and residual norm 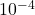 ). Then, we generate a sparse sign embedding of dimension
). Then, we generate a sparse sign embedding of dimension 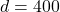 (corresponding to a distortion of roughly
(corresponding to a distortion of roughly 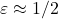 ). Then, we compute the sketch-and-solve solution and, as reference, a “direct” solution by MATLAB’s \.
). Then, we compute the sketch-and-solve solution and, as reference, a “direct” solution by MATLAB’s \.
We compare the quality of the sketch-and-solve solution to the direct solution, using both the residual and forward error:
fprintf('Residuals: sketch-and-solve %.2e, direct %.2e, optimal %.2e\n',...
norm(b-A*sketch_and_solve), norm(b-A*direct), norm(r))
fprintf('Forward errors: sketch-and-solve %.2e, direct %.2e\n',...
norm(x-sketch_and_solve), norm(x-direct))
Here’s the output:
Residuals: sketch-and-solve 1.13e-04, direct 1.00e-04, optimal 1.00e-04
Forward errors: sketch-and-solve 1.06e+03, direct 8.08e-07
The sketch-and-solve solution has a residual norm of 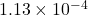 , close to direct method’s residual norm of
, close to direct method’s residual norm of 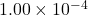 . However, the forward error of sketch-and-solve is
. However, the forward error of sketch-and-solve is 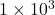 nine orders of magnitude larger than the direct method’s forward error of
nine orders of magnitude larger than the direct method’s forward error of 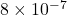 .
.
Does sketch-and-solve work? Ultimately, it’s a question of what kind of accuracy you need for your application. If a small-enough residual is all that’s needed, then sketch-and-solve is perfectly adequate. If small forward error is needed, sketch-and-solve can be quite bad.
One way sketch-and-solve can be improved is by increasing the sketching dimension 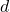 and lowering the distortion
and lowering the distortion  . Unfortunately, improving the distortion of the sketch is expensive. Because of the relation
. Unfortunately, improving the distortion of the sketch is expensive. Because of the relation 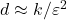 , to decrease the distortion by a factor of ten requires increasing the sketching dimension
, to decrease the distortion by a factor of ten requires increasing the sketching dimension 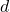 by a factor of one hundred! Thus, sketch-and-solve is really only appropriate when a low degree of distortion
by a factor of one hundred! Thus, sketch-and-solve is really only appropriate when a low degree of distortion  is necessary.
is necessary.
Iterative Sketching: Combining Sketching with Iteration
Sketch-and-solve is a fast way to get a low-accuracy solution to a least-squares problem. But it’s not the only way to use sketching for least-squares. One can also use sketching to obtain high-accuracy solutions by combining sketching with an iterative method.
There are many iterative methods for least-square problems. Iterative methods generate a sequence of approximate solutions 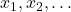 that we hope will converge at a rapid rate to the true least-squares solution,
that we hope will converge at a rapid rate to the true least-squares solution, 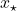 .
.
To using sketching to solve least-squares problems iteratively, we can use the following observation:
If 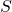 is a sketching matrix for
is a sketching matrix for 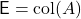 , then
, then 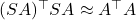 .
.
Therefore, if we compute a QR factorization
![Rendered by QuickLaTeX.com \[SA = QR,\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-9edf84077821e9e0cd0c772b77024799_l3.png)
then
![Rendered by QuickLaTeX.com \[A^\top A \approx (SA)^\top (SA) = R^\top Q^\top Q R = R^\top R.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-90e14ee1172525fd795980cb3ef1a647_l3.png)
Notice that we used the fact that
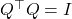
since
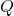
has orthonormal columns. The conclusion is that
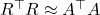
.
Let’s use the approximation 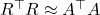 to solve the least-squares problem iteratively. Start off with the normal equations
to solve the least-squares problem iteratively. Start off with the normal equations
(5) ![Rendered by QuickLaTeX.com \[(A^\top A)x = A^\top b. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-3f3c5265345a169d6633a759854510da_l3.png)
We can obtain an approximate solution to the least-squares problem by replacing
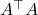
by
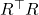
in (5) and solving. The resulting solution is
![Rendered by QuickLaTeX.com \[x_0 = R^{-1} (R^{-\top}(A^\top b)).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-6b63093ea5c1d9714fb0deab43fc4dc3_l3.png)
This solution 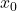 will typically not be a good solution to the least-squares problem (2), so we need to iterate. To do so, we’ll try and solve for the error
will typically not be a good solution to the least-squares problem (2), so we need to iterate. To do so, we’ll try and solve for the error 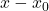 . To derive an equation for the error, subtract
. To derive an equation for the error, subtract 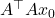 from both sides of the normal equations (5), yielding
from both sides of the normal equations (5), yielding
![Rendered by QuickLaTeX.com \[(A^\top A)(x-x_0) = A^\top (b-Ax_0).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4f1ee993844dbcd8e397f27ccd1deefb_l3.png)
Now, to solve for the error, substitute
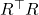
for
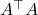
again and solve for
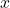
, obtaining a new approximate solution
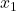
:
![Rendered by QuickLaTeX.com \[x\approx x_1 \coloneqq x_0 + R^{-\top}(R^{-1}(A^\top(b-Ax_0))).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-0496d7e3c8bc117e658c57fc167081be_l3.png)
We can now go another step: Derive an equation for the error 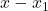 , approximate
, approximate 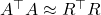 , and obtain a new approximate solution
, and obtain a new approximate solution 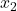 . Continuing this process, we obtain an iteration
. Continuing this process, we obtain an iteration
(6) ![Rendered by QuickLaTeX.com \[x_{i+1} = x_i + R^{-\top}(R^{-1}(A^\top(b-Ax_i))).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f8db238fc978e6a8fc968fc5e387ec27_l3.png)
This iteration is known as the
iterative sketching method. If the distortion is small enough, this method converges at an exponential rate, yielding a high-accuracy least squares solution after a few iterations.
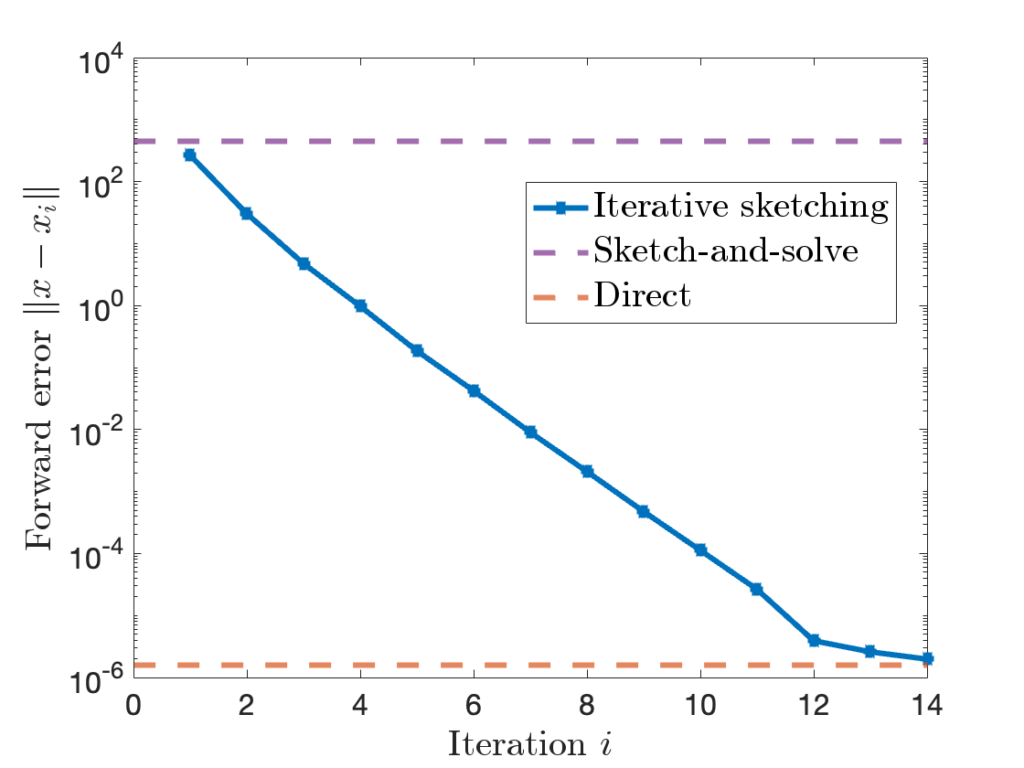
Let’s apply iterative sketching to the example we considered above. We show the forward error of the sketch-and-solve and direct methods as horizontal dashed purple and red lines. Iterative sketching begins at roughly the forward error of sketch-and-solve, with the error decreasing at an exponential rate until it reaches that of the direct method over the course of fourteen iterations. For this problem, at least, iterative sketching gives high-accuracy solutions to the least-squares problem!
To summarize, we’ve now seen two very different ways of using sketching:
- Sketch-and-solve. Sketch the data
 and
and 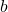 and solve the sketched least-squares problem (3). The resulting solution
and solve the sketched least-squares problem (3). The resulting solution 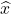 is cheap to obtain, but may have low accuracy.
is cheap to obtain, but may have low accuracy. - Iterative sketching. Sketch the matrix
 and obtain an approximation
and obtain an approximation 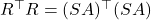 to
to 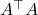 . Use the approximation
. Use the approximation 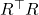 to produce a sequence of better-and-better least-squares solutions
to produce a sequence of better-and-better least-squares solutions 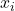 by the iteration (6). If we run for enough iterations
by the iteration (6). If we run for enough iterations 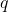 , the accuracy of the iterative sketching solution
, the accuracy of the iterative sketching solution 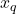 can be quite high.
can be quite high.
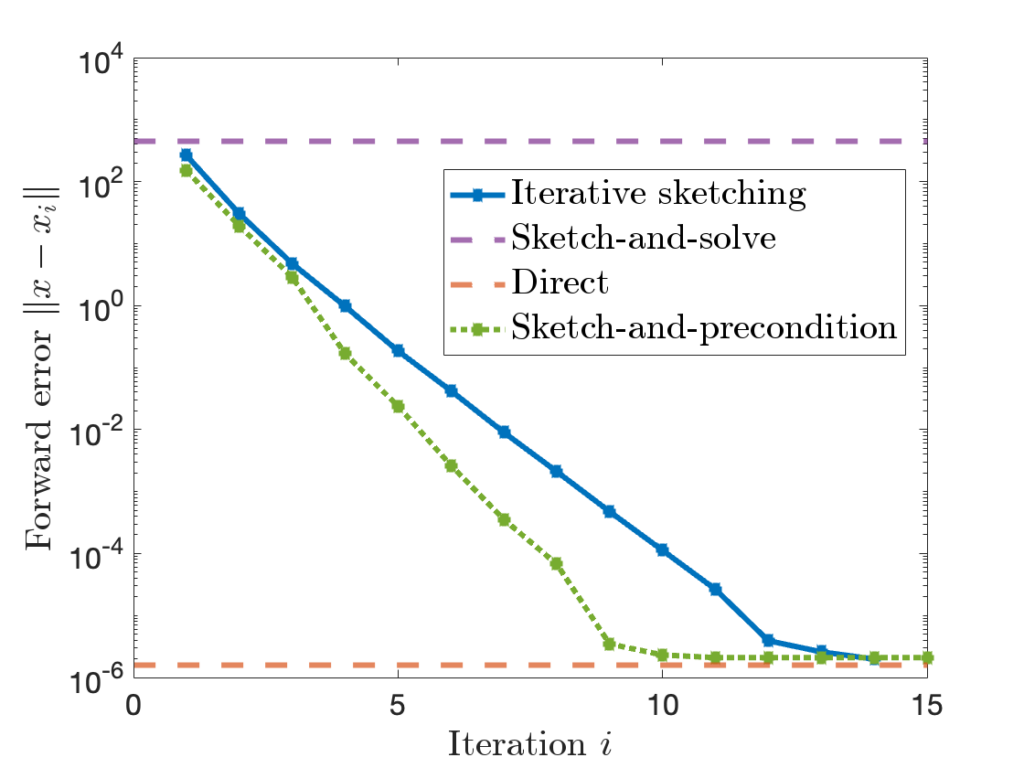
By combining sketching with more sophisticated iterative methods such as conjugate gradient and LSQR, we can get an even faster-converging least-squares algorithm, known as sketch-and-precondition. Here’s the same plot from above with sketch-and-precondition added; we see that sketch-and-precondition converges even faster than iterative sketching does!
“Does sketching work?” Even for a simple problem like least-squares, the answer is complicated:
A direct use of sketching (i.e., sketch-and-solve) leads to a fast, low-accuracy solution to least-squares problems. But sketching can achieve much higher accuracy for least-squares problems by combining sketching with an iterative method (iterative sketching and sketch-and-precondition).
We’ve focused on least-squares problems in this section, but these conclusions could hold more generally. If “sketching doesn’t work” in your application, maybe it would if it was combined with an iterative method.
Just How Accurate Can Sketching Be?
We left our discussion of sketching-plus-iterative-methods in the previous section on a positive note, but there is one last lingering question that remains to be answered. We stated that iterative sketching (and sketch-and-precondition) converge at an exponential rate. But our computers store numbers to only so much precision; in practice, the accuracy of an iterative method has to saturate at some point.
An (iterative) least-squares solver is said to be forward stable if, when run for a sufficient number 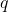 of iterations, the final accuracy
of iterations, the final accuracy  is comparable to accuracy of a standard direct method for the least-squares problem like MATLAB’s \ command or Python’s scipy.linalg.lstsq. Forward stability is not about speed or rate of convergence but about the maximum achievable accuracy.
is comparable to accuracy of a standard direct method for the least-squares problem like MATLAB’s \ command or Python’s scipy.linalg.lstsq. Forward stability is not about speed or rate of convergence but about the maximum achievable accuracy.
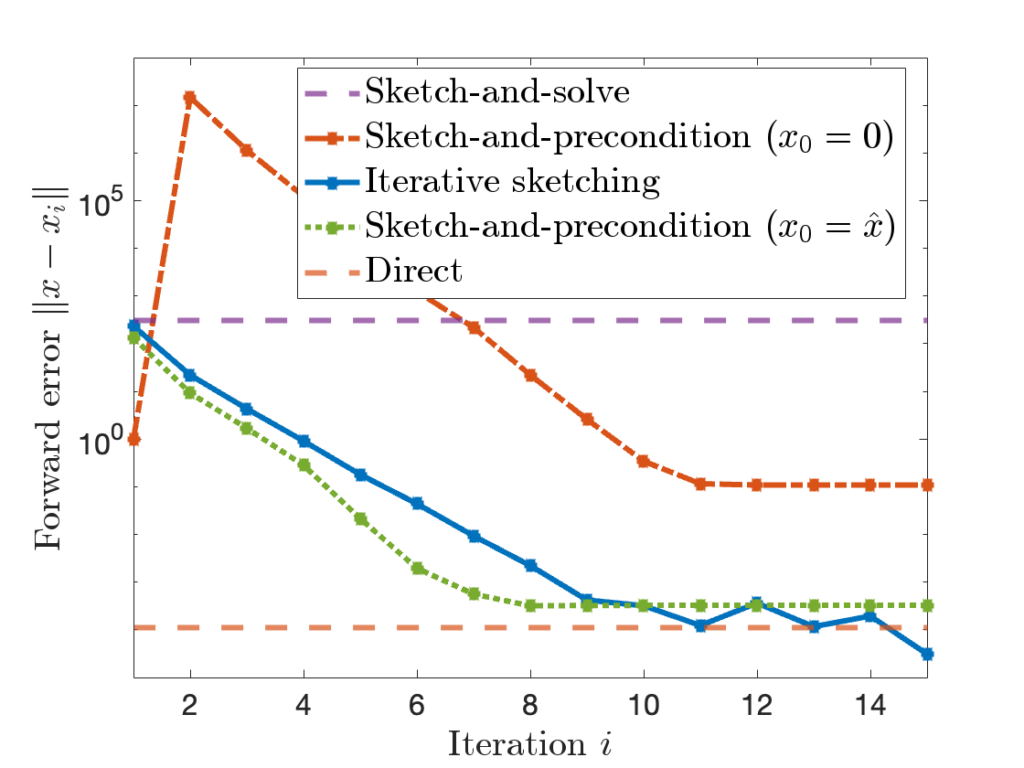
The stability of sketch-and-precondition was studied in a recent paper by Meier, Nakatsukasa, Townsend, and Webb. They demonstrated that, with the initial iterate 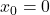 , sketch-and-precondition is not forward stable. The maximum achievable accuracy was worse than standard solvers by orders of magnitude! Maybe sketching doesn’t work after all?
, sketch-and-precondition is not forward stable. The maximum achievable accuracy was worse than standard solvers by orders of magnitude! Maybe sketching doesn’t work after all?
Fortunately, there is good news:
- The iterative sketching method is provably forward stable. This result is shown in my newly released paper; check it out if you’re interested!
- If we use the sketch-and-solve method as the initial iterate
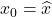 for sketch-and-precondition, then sketch-and-precondition appears to be forward stable in practice. No theoretical analysis supporting this finding is known at present.
for sketch-and-precondition, then sketch-and-precondition appears to be forward stable in practice. No theoretical analysis supporting this finding is known at present.
These conclusions are pretty nuanced. To see what’s going, it can be helpful to look at a graph:
The performance of different methods can be summarized as follows: Sketch-and-solve can have very poor forward error. Sketch-and-precondition with the zero initialization 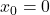 is better, but still much worse than the direct method. Iterative sketching and sketch-and-precondition with
is better, but still much worse than the direct method. Iterative sketching and sketch-and-precondition with 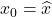 fair much better, eventually achieving an accuracy comparable to the direct method.
fair much better, eventually achieving an accuracy comparable to the direct method.
Put more simply, appropriately implemented, sketching works after all!
Conclusion
Sketching is a computational tool, just like the fast Fourier transform or the randomized SVD. Sketching can be used effectively to solve some problems. But, like any computational tool, sketching is not a silver bullet. Sketching allows you to dimensionality-reduce matrices and vectors, but it distorts them by an appreciable amount. Whether or not this distortion is something you can live with depends on your problem (how much accuracy do you need?) and how you use the sketch (sketch-and-solve or with an iterative method).
![]() be a domain and let
be a domain and let ![]() be a (finite Borel) measure on
be a (finite Borel) measure on ![]() . We consider the task of evaluating
. We consider the task of evaluating ![]()
![]() :
:![]()
![]() . The more smooth
. The more smooth ![]() is, the more accurately we can expect to compute
is, the more accurately we can expect to compute ![]() for a given budget of computational effort.
for a given budget of computational effort.![]() be an RKHS with norm
be an RKHS with norm ![]() . We can interpret the norm as assigning a roughness
. We can interpret the norm as assigning a roughness ![]() to each function
to each function ![]() . If
. If ![]() is large, then
is large, then ![]() is rough; if
is rough; if ![]() is small, then
is small, then ![]() is smooth.
is smooth.![]() is the titular reproducing kernel
is the titular reproducing kernel ![]() . The kernel is a bivariate function
. The kernel is a bivariate function ![]() . It is related to the RKHS inner product
. It is related to the RKHS inner product ![]() by the reproducing property
by the reproducing property![]()
![]() and weights
and weights ![]() . Let’s first assume that the nodes
. Let’s first assume that the nodes ![]() are fixed, and talk about how to pick the weights
are fixed, and talk about how to pick the weights ![]() .
.![]() that we’ll called the ideal weights. There (at least) are five equivalent ways of characterizing the ideal weights. We’ll present all of them. As an exercise, you can try and convince yourself that these characterizations are equivalent, giving rise to the same weights.
that we’ll called the ideal weights. There (at least) are five equivalent ways of characterizing the ideal weights. We’ll present all of them. As an exercise, you can try and convince yourself that these characterizations are equivalent, giving rise to the same weights.![]() .
.![]()
![Rendered by QuickLaTeX.com \[\sum_{j=1}^n k(s_i,s_j)w^\star_j = \int_\Omega k(s_i,x) g(x)\,\mathrm{d}\mu(x) \quad \text{for }i=1,2,\ldots,n.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-1a5ca36966f56dae9d994e208f9ef036_l3.png)
![]() at the nodes, obtaining an interpolant
at the nodes, obtaining an interpolant ![]() . Then, obtain an approximation to the integral by integrating the interpolant:
. Then, obtain an approximation to the integral by integrating the interpolant:![]()
![]()
![]()
![]()
![]() and nodes
and nodes ![]() to be
to be ![]()
![]()
![]() such that
such that ![]() ’s values at any collection of points are (jointly) Gaussian random variables. We write
’s values at any collection of points are (jointly) Gaussian random variables. We write ![]() for a mean-zero Gaussian process with covariance function
for a mean-zero Gaussian process with covariance function ![]() :
: ![]()
![]()
![]()
![]()
![]() . The integral of this random function
. The integral of this random function ![]() is a random variable. To numerically integrate a function
is a random variable. To numerically integrate a function ![]() , compute the expectation of
, compute the expectation of ![]() conditional on
conditional on ![]() agreeing with
agreeing with ![]() at the quadrature nodes:
at the quadrature nodes:![]()
![]() is a probability measure and
is a probability measure and ![]() , it is reasonable to add an additional constraint that the weights
, it is reasonable to add an additional constraint that the weights ![]() lie in the probability simplex
lie in the probability simplex ![Rendered by QuickLaTeX.com \[w\in\Delta\coloneqq \left\{ p\in\real^n_+ : \sum_{i=1}^n p_i = 1\right\}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-55c593b2593abc9ea8c18d4f8edbccd3_l3.png)
![]() ? To pick the nodes, it seems sensible to try and minimize the worst-case error
? To pick the nodes, it seems sensible to try and minimize the worst-case error ![]() with the ideal weights
with the ideal weights ![]() . For this purpose, we can use the following formula:
. For this purpose, we can use the following formula: ![]()
![]()
![]() also suggests a strategy for picking the nodes.
also suggests a strategy for picking the nodes.to make the Nyström approximation
as accurate as possible.
![]() is to
is to ![]() , the smaller the function
, the smaller the function ![]() is and, thus, the smaller the error
is and, thus, the smaller the error ![]()
![]() , it has a high probability of placing the next node
, it has a high probability of placing the next node ![]() far from the previously selected nodes.
far from the previously selected nodes.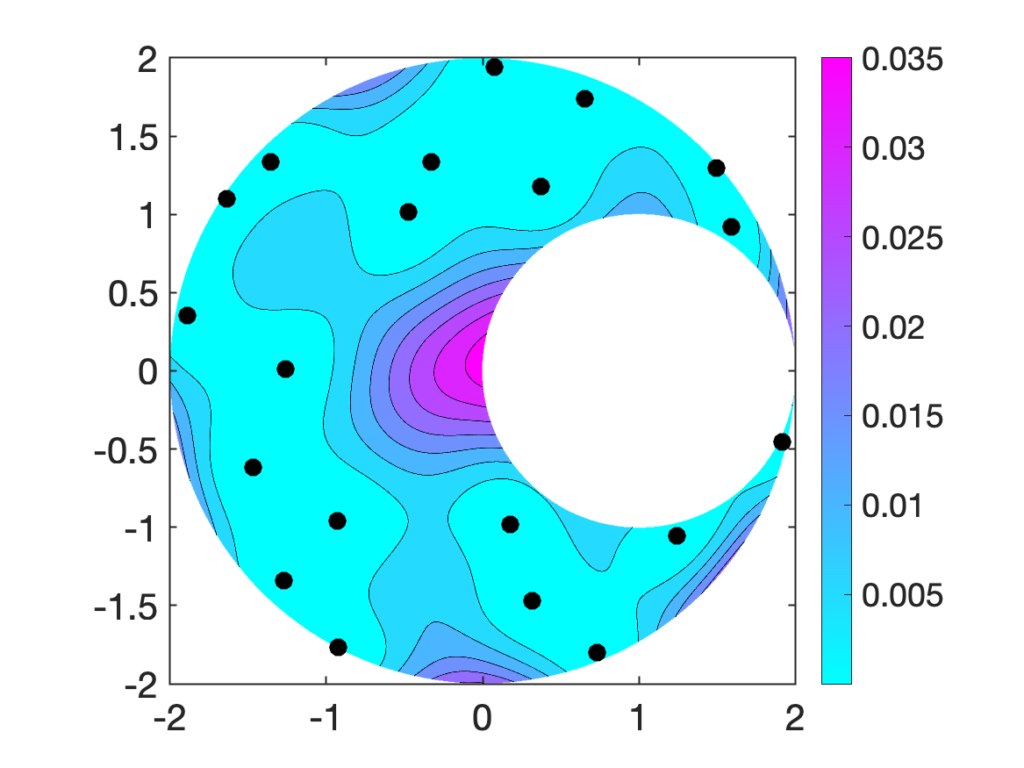
![Rendered by QuickLaTeX.com \[\zeta = \max \left( 8 , \left\lceil 2\sqrt{\frac{d}{k}} \right\rceil \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-5f1ebe7695d9918399b0f97b628fcce1_l3.png)
![Rendered by QuickLaTeX.com \[\prob \{ j_1,\ldots,j_k \text{ are distinct} \} = \prod_{i=1}^k \left(1 - \frac{i-1}{d} \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7dd28281f8dbed8b3a1452a21e7410c8_l3.png)
![Rendered by QuickLaTeX.com \[\mathbb{P} \{ j_1,\ldots,j_k \text{ are distinct} \} \le \prod_{i=0}^{k-1} \exp\left(-\frac{i}{d}\right) = \exp \left( -\frac{1}{d}\sum_{i=0}^{k-1} i \right) = \exp\left(-\frac{k(k-1)}{2d}\right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-8d9d4c7d12af0fd89e53ca261c5df287_l3.png)
![Rendered by QuickLaTeX.com \[\hat{f}_{N}\coloneqq \frac{1}{N}\sum_{n=0}^{N-1} f(x_i).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-591e83a4f5551f6826ddffb5de03ce72_l3.png)
![Rendered by QuickLaTeX.com \[\hat{f}_N = \frac{1}{N}\sum_{n=0}^{N-1} f(x_i)\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-615239005583b665db04e6a578abec08_l3.png)
![Rendered by QuickLaTeX.com \[\langle \varphi_i,\varphi_j \rangle = \expect_{x\sim \pi} [\varphi_i(x)\varphi_j(x)] = \begin{cases}1, & i = j, \\0, & i \ne j.\end{cases}\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-139934705e34440f3c903f0e089f580b_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\expect[f(x_0)f(x_n)] &= \sum_{i=1}^m \expect[f(x_0) f(x_n) \mid x_0 = i] \prob\{x_0 = i\} \\&= \sum_{i=1}^m f(i) \expect[f(x_n) \mid x_0 = i] \pi_i.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-2e350d683f2a7cfff71028de3a45dbf8_l3.png)
![Rendered by QuickLaTeX.com \[\expect[f(x_0)f(x_n)] = \left\langle \sum_{i=1}^m c_i \varphi_i,\sum_{i=1}^m c_i P^n\varphi_i \right\rangle = \left\langle \sum_{i=1}^m c_i \varphi_i,\sum_{i=1}^m c_i \lambda_i^n \varphi_i \right\rangle = \sum_{i=1}^m \lambda_i^n \, c_i^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-63bbc0c0f98d4c57de9a53a7162295f3_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\sigma^2 &= \Var[f(x_0)] + 2\sum_{n=1}^\infty \Cov(f(x_0),f(x_n)) \\&= \sum_{i=2}^m c_i^2 + 2\sum_{n=1}^\infty \sum_{i=2}^m \lambda_i^n \, c_i^2 \\&= -\sum_{i=2}^m c_i^2 + 2\sum_{i=2}^m \left(\sum_{n=0}^\infty \lambda_i^n\right)c_i^2 .\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-da05968b01086497f8e6e098e384b170_l3.png)
![Rendered by QuickLaTeX.com \[\sigma^2 = -\sum_{i=2}^m c_i^2 + 2\sum_{i=2}^m \frac{1}{1-\lambda_i} c_i^2 = \sum_{i=2}^m \left(\frac{2}{1-\lambda_i}-1\right)c_i^2 = \sum_{i=2}^m \frac{1+\lambda_i}{1-\lambda_i} c_i^2. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-61c9cb2ae48b8b474edd697e36a1e58c_l3.png)
![Rendered by QuickLaTeX.com \[\hat{f}_N = \frac{1}{N} \sum_{i=0}^{N-1} f(x_n)\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-76330a2c8101f63831cdc0bcdd4e0cee_l3.png)
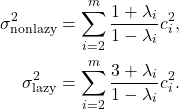
![Rendered by QuickLaTeX.com \[[u_i,u_j]=\begin{cases}1, & i=j, \\0,& i\ne j.\end{cases}\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-6485908bc8e69e0ad6fb16b826344f60_l3.png)
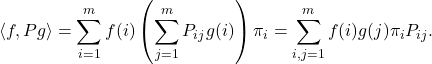
![Rendered by QuickLaTeX.com \[\langle f, Pg \rangle= \sum_{i,j=1}^m f(i) g(j) \pi_jP_{ji} = \sum_{j=1}^m \left( \sum_{i=1}^m f(i) P_{ij} \right) g(j) \pi_j = \langle Pf, g\rangle.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-2e8d0788b8c0f3ee0e4602e817957cfa_l3.png)
![Rendered by QuickLaTeX.com \[\langle\varphi_i,\varphi_j\rangle=\begin{cases}1, &i=j\\0,& i\ne j.\end{cases}\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-fd0e99bc2439329beccce1339db0aee9_l3.png)
![Rendered by QuickLaTeX.com \[\chi^2(\sigma \mid\mid \pi) \coloneqq \Var \left(\frac{d\sigma}{d\pi} \right) = \expect \left[\left( \frac{d\sigma}{d\pi} - 1 \right)^2\right] = \expect \left[\left(\frac{d\sigma}{d\pi}\right)^2\right] - 1.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-2521b63ae50f02fe6099c54dcc4e00a4_l3.png)
![Rendered by QuickLaTeX.com \[\norm{\sigma - \pi}_{\rm TV} = \frac{1}{2} \expect \left[ \left| \frac{d\sigma}{d\pi} - 1 \right| \right] \le \frac{1}{2} \left(\expect \left[ \left( \frac{d\sigma}{d\pi} - 1 \right)^2 \right]\right)^{1/2} = \frac{1}{2} \sqrt{\chi^2(\sigma \mid\mid \pi)}. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4986b7434b84b79aff6f94df2ec4b4e8_l3.png)
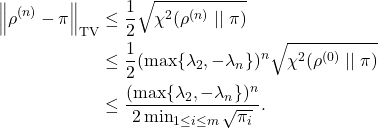
![Rendered by QuickLaTeX.com \[\left( \frac{d\rho^{(n+1)}}{d\pi}\right)_j = \frac{\rho_j^{(n+1)}}{\pi_j} = \frac{\sum_{i=1}^m \rho^{(n)}_i P_{ij}}{\pi_j}= \sum_{i=1}^m \rho^{(n)}_i\frac{P_{ij}}{\pi_j}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-b6c60937b23d2509e4a3e4fa1f121427_l3.png)
![Rendered by QuickLaTeX.com \[\left( \frac{d\rho^{(n+1)}}{d\pi}\right)_j = \sum_{i=1}^m\rho^{(n)}_i \frac{P_{ji}}{\pi_i} = \sum_{i=1}^m P_{ji} \frac{\rho^{(n)}_i}{\pi_i} = \left( P \frac{d \rho^{(n)}}{d\pi} \right)_j.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d22315b80e3fc4d3736d0c49f877fa9c_l3.png)
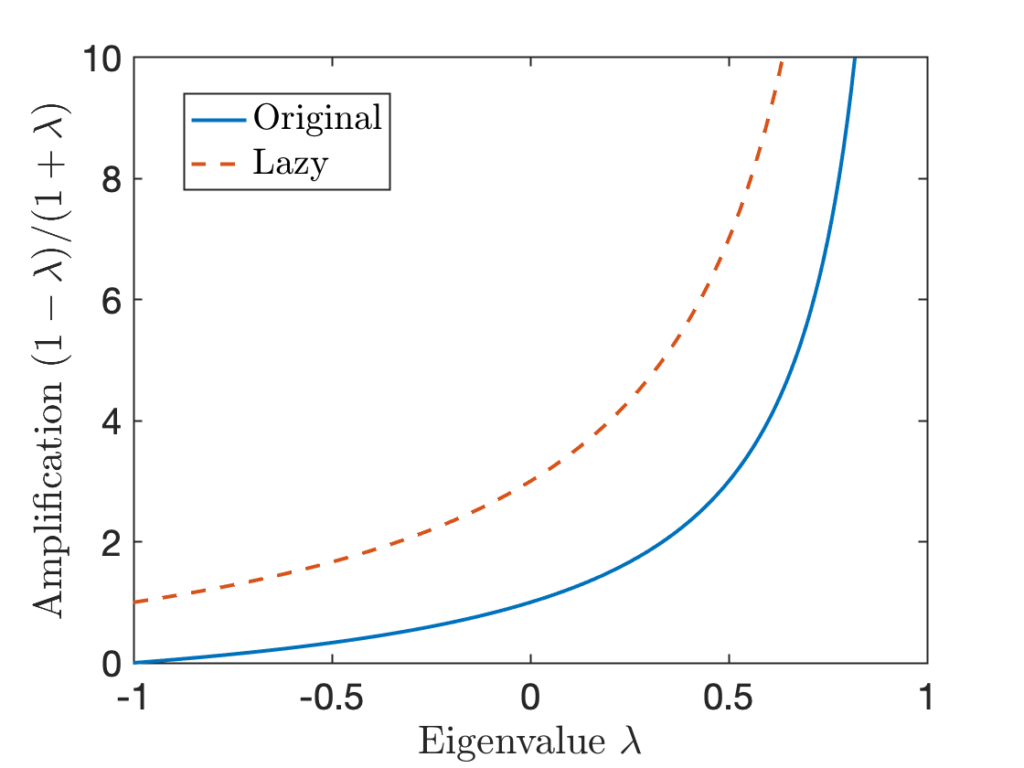
![Rendered by QuickLaTeX.com \[\chi^2(\rho^{(n)} \mid\mid \pi) = \sum_{i=2}^m c_i^2 \lambda_i^{2n} \le (\max \{ \lambda_2, -\lambda_m \})^{2n} \sum_{i=2}^m c_i^2 = (\max \{ \lambda_2, -\lambda_m \})^{2n} \chi^2(\rho^{(0)} \mid\mid \pi).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7b82e2d3cefcede8d4a7a72b1fa3ceea_l3.png)
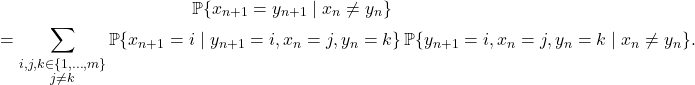
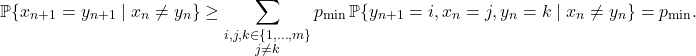
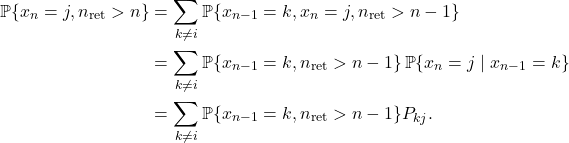
![Rendered by QuickLaTeX.com \[a_j = P_{ij} + \sum_{n=2}^\infty \sum_{k\ne i} \prob\{x_{n-1} = k, n_{\rm ret} > n-1 \} P_{kj}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f243b52714b96a419b94980a12f078ee_l3.png)
![Rendered by QuickLaTeX.com \[a_j = P_{ij} + \sum_{k\ne i} \left(\sum_{n=1}^\infty \prob\{x_n = k, n_{\rm ret} > n \}\right) P_{kj}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-cad1677f621e66a91f850ab352cedce0_l3.png)
![Rendered by QuickLaTeX.com \[a_j = a_iP_{ij} + \sum_{k\ne i} a_k P_{kj} = \sum_{k=1}^m a_k P_{kj},\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-cecaa5c06b9ce9d9c3f4aa814dfc2323_l3.png)
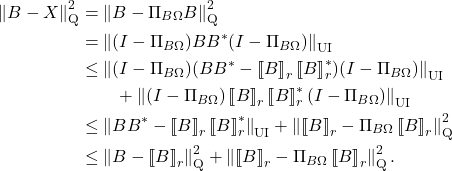
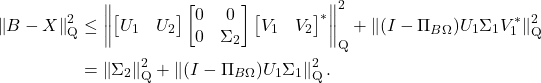
![Rendered by QuickLaTeX.com \begin{align*}\Pi_{B\Omega\Omega_1^\dagger} &= (B\Omega\Omega_1^\dagger) \left[(B\Omega\Omega_1^\dagger)^*(B\Omega\Omega_1^\dagger)\right]^\dagger (B\Omega\Omega_1^\dagger)^* \\&= (U_1\Sigma_1 + U_2\Sigma_2\Omega_2\Omega_1^\dagger) \left[\Sigma_1^2 + (\Sigma_2\Omega_2\Omega_1^\dagger)^*(\Sigma_2\Omega_2\Omega_1^\dagger)\right]^\dagger (U_1\Sigma_1 + U_2\Sigma_2\Omega_2\Omega_1^\dagger)^*.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d9e0cd585357787c460f6471f52ecb0a_l3.png)
![Rendered by QuickLaTeX.com \[a:h \coloneqq \lim_{b\downarrow a, \: k\downarrow h} b:k = \begin{cases}\left( a^{-1}+h^{-1}\right)^{-1}, & a,h > 0 ,\\0, & \textrm{otherwise}.\end{cases}\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7c49cbcbcd8d8f75fa4f18a40562e718_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\left\|A:H\right\|_{\rm UI} &= \max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr((A:H)M) \\&= \max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr(M^{1/2}(A:H)M^{1/2})\\&= \max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr((M^{1/2}AM^{1/2}):(M^{1/2}HM^{1/2})) \\&\le \max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \left[\tr(M^{1/2}AM^{1/2}):\tr(M^{1/2}HM^{1/2}) \right] \\&\le \left[\max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr(M^{1/2}AM^{1/2})\right]:\left[\max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr(M^{1/2}HM^{1/2})\right]\\&= \left\|A\right\|_{\rm UI} : \left\|H\right\|_{\rm UI}.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4d5c2226a0ddf81d346da401ff4b53b9_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\left\|B - X\right\|_{\rm Q}^2&\le \left\| \Sigma_2 \right\|_{\rm Q}^2 + \left\|\Sigma_1^2 : [(\Sigma_2\Omega_2\Omega_1^\dagger)^*(\Sigma_2\Omega_2\Omega_1^\dagger)]\right\|_{\rm UI} \\&\le \left\| \Sigma_2 \right\|_{\rm Q}^2 + \left\|\Sigma_1^2\right\|_{\rm UI} : \left\|[(\Sigma_2\Omega_2\Omega_1^\dagger)^*(\Sigma_2\Omega_2\Omega_1^\dagger)]\right\|_{\rm UI} \\&= \left\| \Sigma_2 \right\|_{\rm Q}^2 + \left\|\Sigma_1\right\|_{\rm Q}^2 : \left\|\Sigma_2\Omega_2\Omega_1^\dagger\right\|^2_{\rm Q}.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-df07b0b9660af51129fd29552ad6c650_l3.png)
![Rendered by QuickLaTeX.com \[\mathbb{E} \left\|SGW\right\|_{\rm F}^2 = \mathbb{E} \sum_{ij} \left(\sum_{k\ell} s_{ik}g_{k\ell}w_{\ell j} \right)^2 = \mathbb{E} \sum_{ijk\ell} s_{ik}^2 w_{\ell j}^2 = \left\|S\right\|_{\rm F}^2\left\|W\right\|_{\rm F}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-dc219fd0dab2748965ecbf439c051db5_l3.png)
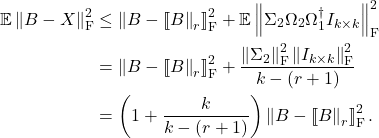
![Rendered by QuickLaTeX.com \[\mathbb{E} \left\|B - X\right\|^2 \le \min_{r \le k-2} \left( 1 + \frac{2r}{k-(r+1)} \right) \left(\sigma_{r+1}^2 + \frac{\mathrm{e}^2}{k-r} \sum_{i>r} \sigma_i^2 \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d4bb8b98ee0928e647c7cdbd91332fcc_l3.png)
![Rendered by QuickLaTeX.com \[\mathbb{E}[ f(X_t+\Delta)]=\mathbb{E}[f(X_t)] + \mathbb{E}[f'(X_t)\Delta] + \frac{1}{2} \mathbb{E}[f''(X_t)] \mathbb{E}[\Delta^2] + \underbrace{\frac{1}{6} \mathbb{E}[f'''(\xi) \Delta^3]}_{:=\mathrm{Rem}(\delta)}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-ccacfa2622a31446e66624a9a8458dc4_l3.png)