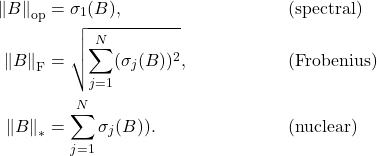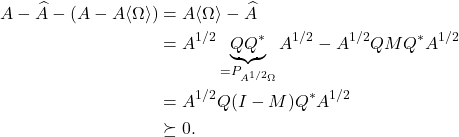I am delighted to share that my paper Randomly pivoted Cholesky: Randomly pivoted Cholesky: Practical approximation of a kernel matrix with few entry evaluations, joint with Yifan Chen, Joel Tropp, and Robert Webber, has been published online in Communications on Pure and Applied Mathematics. To celebrate this occasion, I want to share one of my favorite tricks in the design of low-rank approximation algorithms, which I will call the Gram correspondence.
Projection Approximation and Nyström Approximation
When we construct a low-rank approximation to a matrix, the type of approximation we use is typically dictated by the size and properties of the matrix. For a rectangular matrix ![]() , one of the standard techniques is the standard randomized SVD algorithm:
, one of the standard techniques is the standard randomized SVD algorithm:
- Generate a Gaussian random matrix
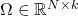 .
. - Form the product
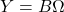 .
. - Compute an (economy-size) QR decomposition
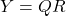 .
. - Evaluate the SVD
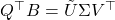 .
. - Output the low-rank approximation
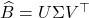 for
for 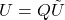 .
.
Succinctly, the output of the randomized SVD is given by the formula ![]() , where
, where ![]() denotes the orthogonal projector onto the column span of a matrix
denotes the orthogonal projector onto the column span of a matrix ![]() . This motivates the general definition:
. This motivates the general definition:
Definition (projection approximation): Given a test matrix
, the projection approximation to the matrix
is
.
The class of projection approximations is much richer than merely the approximation constructed by the standard randomized SVD. Indeed, low-rank approximations computed by randomized subspace iteration, randomized block Krylov iteration, column-pivoted QR decompositions, etc. all fit under the umbrella of projection approximations.
Many matrices in applications have additional structure such as symmetry or sparsity, and it can be valuable to make use of low-rank approximations that take advantage of those properties. An especially important type of structure is positive semidefiniteness. For our purposes, a positive semidefinite matrix ![]() is one that is symmetric and possesses nonnegative eigenvalues, and we will use abbreviate “positive semidefinite” as “psd”. Psd matrices arise in applications as covariance matrices, kernel matrices, and as discretizations of certain differential and integral operators. Further, any rectangular matrix
is one that is symmetric and possesses nonnegative eigenvalues, and we will use abbreviate “positive semidefinite” as “psd”. Psd matrices arise in applications as covariance matrices, kernel matrices, and as discretizations of certain differential and integral operators. Further, any rectangular matrix ![]() gives rise to its psd Gram matrix
gives rise to its psd Gram matrix ![]() ; we will have much more to say about Gram matrices below.
; we will have much more to say about Gram matrices below.
To compute a low-rank approximation to a psd matrix, the preferred format as usually the Nyström approximation:
Definition (Nyström approximation): Given a test matrix
, the Nyström approximation is1Throughout this post, the inverse
is interpreted as the Moore–Penrose pseudoinverse if its argument is not invertible.
As discussed in a previous post, the general class of Nyström approximations includes many useful specializations depending on how the matrix ![]() is selected.
is selected.
The Gram Correspondence
The Gram correspondence is a connection between projection approximation and Nyström approximation:
The Gram correspondence: Let
be any rectangular matrix and consider the Gram matrix
. Fix a test matrix
, and define the Nyström approximation
to
and the projection approximation
to
. Then
That is, the Gram matrix
of the projection approximation
is the Nyström approximation
.
As we will see, this correspondence has many implications:
- Special cases. The correspondence contains several important facts as special cases. Examples include the equivalence between the randomized SVD and single-pass Nyström approximation and the equivalence of (partial) column-pivoted QR factorization and (partial) pivoted Cholesky decomposition.
- Algorithm design. Since Nyström approximation and projection approximations are closely related, one can often interconvert algorithms for one type of approximation into the other. These conversions can lead one to discover new algorithms. We will provide a historical answer with the discovery of randomly pivoted Cholesky.
- Error bounds. The Gram correspondence is immensely helpful in the analysis of algorithms, as it makes error bounds for projection approximations and Nyström approximations easily derivable from each other.
For those interested, we give a short proof of the Gram correspondence below.
Aside: Gram Square Roots
Before we go forward, let me highlight a point of potential conclusion and introduce some helpful terminology. When thinking about algorithms for a psd matrix ![]() , it will often to be helpful to conjure a matrix
, it will often to be helpful to conjure a matrix ![]() for which
for which ![]() . Given a psd matrix
. Given a psd matrix ![]() , there is always a matrix
, there is always a matrix ![]() for which
for which ![]() , but this
, but this ![]() is not unique. Indeed, if
is not unique. Indeed, if ![]() , then we have the infinite family of decompositions
, then we have the infinite family of decompositions ![]() generated by every matrix
generated by every matrix ![]() with orthonormal columns. This motivates the following definition:
with orthonormal columns. This motivates the following definition:
Definition (Gram square root): A matrix
is a Gram square root for
if
.
A Gram square root ![]() for
for ![]() need not be a square root in the traditional sense that
need not be a square root in the traditional sense that ![]() . Indeed, a Gram square root
. Indeed, a Gram square root ![]() can be rectangular, so
can be rectangular, so ![]() need not even be defined.2The Gram square root should be contrasted with a different type of square root, the matrix square root written
need not even be defined.2The Gram square root should be contrasted with a different type of square root, the matrix square root written ![]() . The matrix square root
. The matrix square root ![]() is square and psd, and satisfies
is square and psd, and satisfies ![]() . Moreover, it is the unique matrix with these properties. Using the Gram square root terminology, the Gram correspondence can be written more succinctly:
. Moreover, it is the unique matrix with these properties. Using the Gram square root terminology, the Gram correspondence can be written more succinctly:
Gram correspondence (concise): If
is any Gram square root of
, then the projection approximation
is a Gram square root of the Nyström approximation
.
Examples of the Gram Correspondence
Before going further, let us see a couple of explicit examples of the Gram correspondence.
Randomized SVD and Single-Pass Nyström Approximation
The canonical example of the Gram correspondence is the equivalence of the randomized SVD algorithm and the single-pass Nyström approximation. Let ![]() be a Gram square root for a psd matrix
be a Gram square root for a psd matrix ![]() . The randomized SVD of
. The randomized SVD of ![]() is given by the following steps, which we saw in the introduction:
is given by the following steps, which we saw in the introduction:
- Generate a Gaussian random matrix
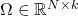 .
. - Form the product
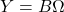 .
. - Compute a QR decomposition
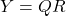 .
. - Evaluate the SVD
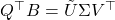 .
. - Output the low-rank approximation
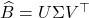 for
for 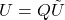 .
.
Now, imagine taking the same Gaussian matrix ![]() from the randomized SVD, and use it to compute the Nyström approximation
from the randomized SVD, and use it to compute the Nyström approximation ![]() . Notice that this Nyström approximation can be computed in a single pass over the matrix
. Notice that this Nyström approximation can be computed in a single pass over the matrix ![]() . Namely, use a single pass over
. Namely, use a single pass over ![]() to compute
to compute ![]() and use following formula:3In practice, one should be careful about implementation for a single-pass Nyström approximation; see this paper for details.
and use following formula:3In practice, one should be careful about implementation for a single-pass Nyström approximation; see this paper for details.
![]()
The Gram correspondence tells us that the randomized SVD and single-pass Nyström approximation are closely related, in the sense that ![]() . The randomized SVD approximation
. The randomized SVD approximation ![]() is a Gram square root of the single-pass Nyström approximation
is a Gram square root of the single-pass Nyström approximation ![]() .
.
Column-Pivoted QR Factorization and Pivoted Cholesky Decomposition
A more surprising consequence of the Gram correspondence is the connection between low-rank approximations produced by partial column-pivoted QR factorization of a rectangular matrix ![]() and a partial pivoted Cholesky decomposition of
and a partial pivoted Cholesky decomposition of ![]() . Let’s begin by describing these two approximation methods.
. Let’s begin by describing these two approximation methods.
Let’s begin with pivoted partial Cholesky decomposition. Let ![]() be a psd matrix, and initialize the zero approximation
be a psd matrix, and initialize the zero approximation ![]() . For
. For ![]() , perform the following steps:
, perform the following steps:
- Choose a column index
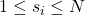 . These indices are referred to as pivot indices or, more simply, pivots.
. These indices are referred to as pivot indices or, more simply, pivots. - Update the low-rank approximation
![Rendered by QuickLaTeX.com \[\hat{A} \gets \hat{A} + \frac{A(:,s_i)A(s_i,:)}{A(s_i,s_i)}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-ec07289a72875184db6bb7476af3149e_l3.png)
- Update the matrix
![Rendered by QuickLaTeX.com \[A \gets A - \frac{A(:,s_i)A(s_i,:)}{A(s_i,s_i)}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-0f9632b22fb854d1c90533858afc4321_l3.png)
Here, we are using MATLAB notation to index the matrix ![]() . The output of this procedure is
. The output of this procedure is
![]()
Given the Gram correspondence, we expect that this pivoted partial Cholesky procedure for computing a Nyström approximation to a psd matrix ![]() should have an analog for computing a projection approximation to a rectangular matrix
should have an analog for computing a projection approximation to a rectangular matrix ![]() . This analog is given by the partial column-pivoted QR factorization, which produces a low-rank approximation according as follows. Let
. This analog is given by the partial column-pivoted QR factorization, which produces a low-rank approximation according as follows. Let ![]() be a psd matrix, and initialize the zero approximation
be a psd matrix, and initialize the zero approximation ![]() . For
. For ![]() , perform the following steps:
, perform the following steps:
- Choose a pivot index
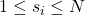 .
. - Update the low-rank approximation by adding the projection of
 onto the selected column:
onto the selected column: 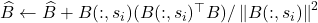 .
. - Update the matrix by removing the projection of
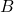 onto the selected column:
onto the selected column: 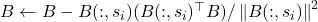 .
.
The output of this procedure is the column projection approximation ![]() , which is an example of the general projection approximation with
, which is an example of the general projection approximation with ![]() .4The column projection approximation is often presented in factorized form in one of two ways. First,
.4The column projection approximation is often presented in factorized form in one of two ways. First, ![]() can be expressed as
can be expressed as ![]() , where
, where ![]() is a matrix with orthonormal columns and
is a matrix with orthonormal columns and ![]() is an upper trapezoidal matrix, up to a permutation of the rows. This factorized form is easy to compute roughly following steps 1–3 above, which explains why we call that procedure a “QR factorization“. Second,
is an upper trapezoidal matrix, up to a permutation of the rows. This factorized form is easy to compute roughly following steps 1–3 above, which explains why we call that procedure a “QR factorization“. Second, ![]() can be factorized
can be factorized ![]() for a weight matrix
for a weight matrix ![]() . This type of factorization is known as an interpolative decomposition.
. This type of factorization is known as an interpolative decomposition.
The pivoted partial Cholesky and QR factorizations are very traditional ways of computing a low-rank approximation in numerical linear algebra. The Gram correspondence tells us immediately that these two approaches are closely related:
Let
be a Gram square root of
. Compute a column projection approximation
to
using a partially column-pivoted QR factorization with pivots
, and compute a column Nyström approximation
to
using a partial pivoted Cholesky decomposition with the same set of pivots
. Then
.
I find this remarkable: the equivalence of the randomized SVD and single-pass randomized Nyström approximation and the equivalence of (partial pivoted) Cholesky and QR factorizations are both consequences of the same general principle, the Gram correspondence.
Using the Gram Correspondence to Discover Algorithms
The Gram correspondence is more than just an interesting way of connecting existing types of low-rank approximations: It can be used to discover new algorithms. We can illustrate this with an example, the randomly pivoted Cholesky algorithm.
Background: The Randomly Pivoted QR Algorithm
In 2006, Deshpande, Rademacher, Vempala, and Wang discovered an algorithm that they called adaptive sampling for computing a projection approximation to a rectangular matrix ![]() . Beginning from the trivial initial approximation
. Beginning from the trivial initial approximation ![]() , algorithm proceeds as follows for
, algorithm proceeds as follows for ![]() :
:
- Choose a column index
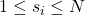 randomly according to the squared column norm distribution:
randomly according to the squared column norm distribution: ![Rendered by QuickLaTeX.com \[\prob\{s_i = j\} = \frac{\norm{B(:,j)}^2}{\norm{B}_{\rm F}^2} \quad \text{for } j=1,2,\ldots,k.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-9dd2bd2b1a2336439460e2d9edc93ac1_l3.png)
- Update the low-rank approximation by adding the projection of
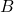 onto the selected column:
onto the selected column: 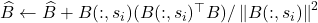 .
. - Update the matrix by removing the projection of
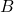 onto the selected column:
onto the selected column: 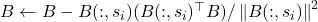 .
.
Given our discussion above, we recognize this as an example of partial column-pivoted QR factorization with a particular randomized procedure for selecting the pivots ![]() .5Interestingly, Deshpande and coauthors did not make the connection between their approach and pivoted QR algorithms in their work. Therefore, it can also be convenient to call this algorithm randomly pivoted QR.
.5Interestingly, Deshpande and coauthors did not make the connection between their approach and pivoted QR algorithms in their work. Therefore, it can also be convenient to call this algorithm randomly pivoted QR.
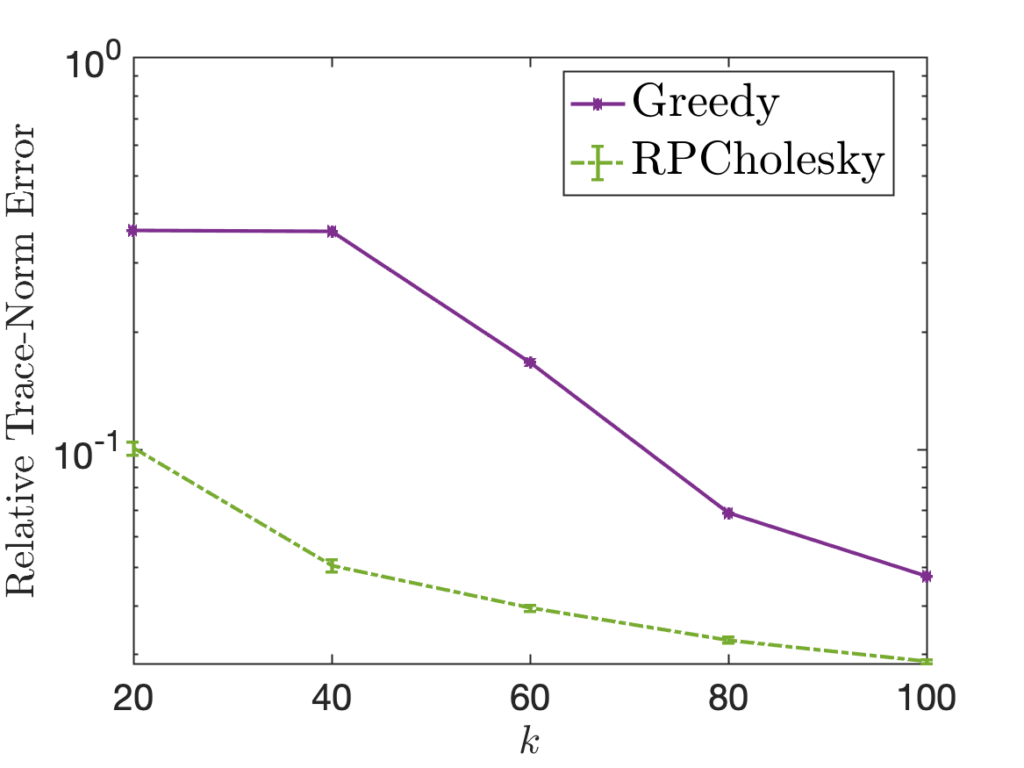
Randomly pivoted QR is a nice algorithm for rectangular low-rank approximation. Each step requires a full pass over the matrix and ![]() operations, so the full procedure requires
operations, so the full procedure requires ![]() operations. This makes the cost of the algorithm similar to other methods for rectangular low-rank approximation such as the randomized SVD, but it has the advantage that it computes a column projection approximation.6There are other algorithms for computing a high-quality column projection approximation in
operations. This makes the cost of the algorithm similar to other methods for rectangular low-rank approximation such as the randomized SVD, but it has the advantage that it computes a column projection approximation.6There are other algorithms for computing a high-quality column projection approximation in ![]() operations, such as sketchy pivoting. Variants of these approaches are compared in this recent paper. However, randomly pivoted QR is not a particularly effective algorithm for computing a low-rank approximation to a psd matrix, since—as we shall see—there are faster procedures available.
operations, such as sketchy pivoting. Variants of these approaches are compared in this recent paper. However, randomly pivoted QR is not a particularly effective algorithm for computing a low-rank approximation to a psd matrix, since—as we shall see—there are faster procedures available.
Following the Gram correspondence, we expect there should be an analog of the randomly pivoted QR algorithm for computing a low-rank approximation of a psd matrix. That algorithm, which we call randomly pivoted Cholesky, is derived from randomly pivoted QR in the following optional section:
Randomly Pivoted Cholesky
That derivation was a bit complicated, so let’s summarize. We can with the randomly pivoted QR algorithm for computing a projection approximation to a rectangular matrix ![]() , and we used it to derive a randomly pivoted Cholesky algorithm for computing a column Nyström approximation to a psd matrix
, and we used it to derive a randomly pivoted Cholesky algorithm for computing a column Nyström approximation to a psd matrix ![]() . Removing the cruft of the derivation, this algorithm is very simple to state:
. Removing the cruft of the derivation, this algorithm is very simple to state:
- Choose a column index
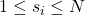 randomly according to the diagonal distribution:
randomly according to the diagonal distribution: ![Rendered by QuickLaTeX.com \[\prob\{s_i = j\} = \frac{A(j,j)}{\tr(A)} \quad \text{for } j=1,2,\ldots,k.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-750f3165cab9fdf250259d6e294fabfc_l3.png)
- Update the low-rank approximation
![Rendered by QuickLaTeX.com \[\hat{A} \gets \hat{A} + \frac{A(:,s_i)A(s_i,:)}{A(s_i,s_i)}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-ec07289a72875184db6bb7476af3149e_l3.png)
- Update the matrix
![Rendered by QuickLaTeX.com \[A \gets A - \frac{A(:,s_i)A(s_i,:)}{A(s_i,s_i)}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-0f9632b22fb854d1c90533858afc4321_l3.png)
I find this to be remarkably cool. We started with a neat algorithm (randomly pivoted QR) for approximating rectangular matrices, and we used the Gram correspondence to derive a different randomly pivoted Cholesky algorithm for psd matrix approximation!
And randomly pivoted Cholesky allows us to pull a cool trick that we couldn’t with randomly pivoted QR. Observe that the randomly pivoted Cholesky algorithm only ever interacts with the residual matrix ![]() through the selected pivot columns
through the selected pivot columns ![]() and through the diagonal entries
and through the diagonal entries ![]() . Therefore, we can derive an optimized version of the randomly pivoted Cholesky algorithm that only reads
. Therefore, we can derive an optimized version of the randomly pivoted Cholesky algorithm that only reads ![]() entries of the matrix
entries of the matrix ![]() (
(![]() columns plus the diagonal) and requires only
columns plus the diagonal) and requires only ![]() operations! See our paper for details.
operations! See our paper for details.
So we started with the randomly pivoted QR algorithm, which requires ![]() operations, and we used it to derive the randomly pivoted Cholesky algorithm that runs in
operations, and we used it to derive the randomly pivoted Cholesky algorithm that runs in ![]() operations. Let’s make this concrete with some specific numbers. Setting
operations. Let’s make this concrete with some specific numbers. Setting ![]() and
and ![]() , randomly pivoted QR requires roughly
, randomly pivoted QR requires roughly ![]() (100 trillion) operations and randomly pivoted Cholesky requires roughly
(100 trillion) operations and randomly pivoted Cholesky requires roughly ![]() (10 billion) operations, a factor of 10,000 smaller operation count!
(10 billion) operations, a factor of 10,000 smaller operation count!
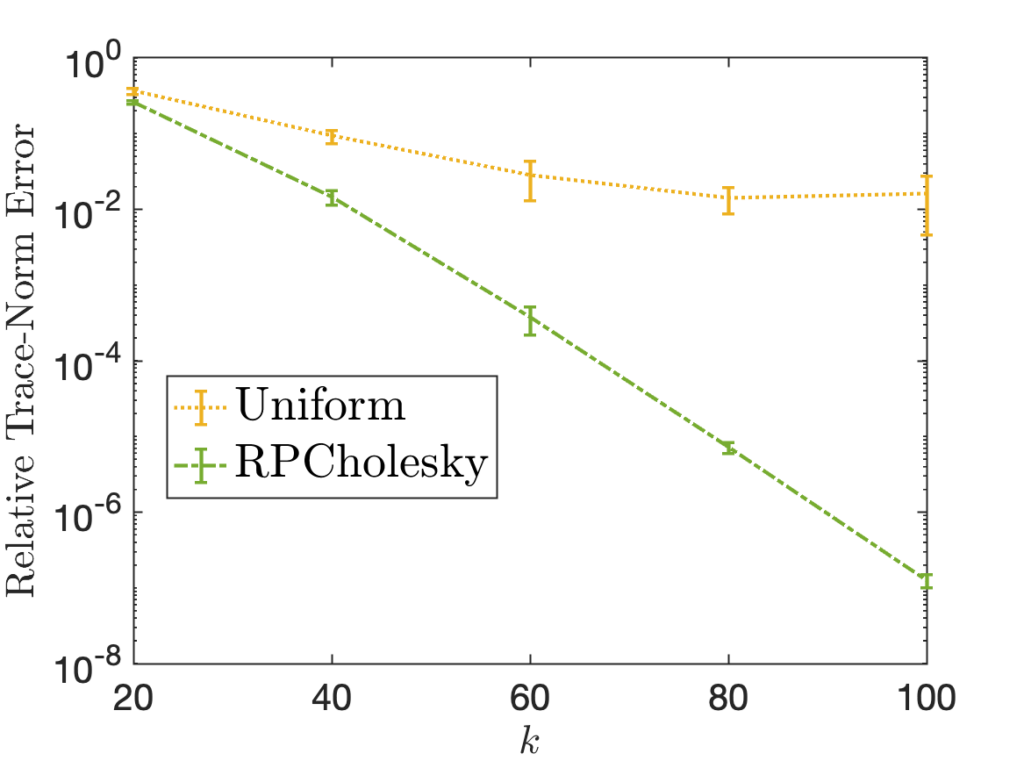
Randomly pivoted Cholesky has an interesting history. It appears to have first appeared in print in the work of Musco and Woodruff (2017), who used the Gram correspondence to derive the algorithm from randomly pivoted QR. It is remarkable that it took a full 11 years after Deshpande and co-author’s original work on randomly pivoted QR for randomly pivoted Cholesky to be discovered. Even after Musco and Woodruff’s paper, the algorithm appears to have largely been overlooked for computation in practice, and I am unaware of any paper documenting computational experiments with randomly pivoted Cholesky before our paper was initially released in 2022.8A partial exception to this statement is the work of Poulson (2020), who used randomly pivoted Cholesky to sample from determinantal point processes. Poulson’s setting is quite different from ours: his input ![]() is always a rank-
is always a rank-![]() orthogonal projection matrix, he runs for exactly
orthogonal projection matrix, he runs for exactly ![]() steps, and he uses the pivot set
steps, and he uses the pivot set ![]() as output. Our paper reexamined the randomly pivoted Cholesky procedure, providing computational experiments comparing it to alternatives and using it for scientific machine learning applications, and provided new error bounds.
as output. Our paper reexamined the randomly pivoted Cholesky procedure, providing computational experiments comparing it to alternatives and using it for scientific machine learning applications, and provided new error bounds.
Other Examples of Algorithm Design by the Gram Correspondence
The Gram correspondence gives a powerful tool for inventing new algorithms or showing equivalence between existing algorithms. There are many additional examples, such as block (and rejection sampling-accelerated) versions of randomly pivoted QR/Cholesky, Nyström versions of randomized block Krylov iteration, and column projection/Nyström approximations generated by determinantal point process sampling. Indeed, if you invent a new low-rank approximation algorithm, it is always worth checking: Using the Gram correspondence, can you get another algorithm for free?
Transference of Error Bounds
The Gram correspondence also allows us to transfer error bounds between projection approximations and Nyström approximations. Let’s begin with the simplest manifestation of this principle:
Transference of Error Bounds I: Let
be a Gram square root of
, and let
and
be the corresponding projection and Nyström approximations generated using the same test matrix
. Then
This shows that the Frobenius norm error of a projection approximation and the trace error of a Nyström approximation are the same.9Note that, for any Nyström approximation ![]() to
to ![]() , the residual
, the residual ![]() is psd. Therefore, the trace error
is psd. Therefore, the trace error ![]() is nonnegative and satisfies
is nonnegative and satisfies ![]() if and only if
if and only if ![]() ; these statements justify why
; these statements justify why ![]() is an appropriate expression for measuring the error of the approximation
is an appropriate expression for measuring the error of the approximation ![]() . Using this result, we can immediately transfer error bounds for projection approximations to Nyström approximations and visa versa. For instance, in this previous post, we proved the following error bound for the rank-
. Using this result, we can immediately transfer error bounds for projection approximations to Nyström approximations and visa versa. For instance, in this previous post, we proved the following error bound for the rank-![]() randomized SVD (with a Gaussian test matrix
randomized SVD (with a Gaussian test matrix ![]() ):
):
![]()
![]()
For those interested, one can extend the transference of error bounds to general unitarily invariant norms. See the following optional section for details:
Conclusion
This has been a long post about a simple idea. Many of the low-rank approximation algorithms in the literature output a special type of approximation, either a projection approximation or a Nyström approximation. As this post has shown, these two types of approximation are equivalent, with algorithms and error bounds for one type of approximation transferring immediately to the other format. This equivalence is a powerful tool for the algorithm designer, allowing us to discover new algorithms from old ones, like randomly pivoted Cholesky from randomly pivoted QR.
![Rendered by QuickLaTeX.com \[\hat{A} &\gets \left(\hat{B} + \frac{B(:,s_i) (B(:,s_i)^\top B)}{\norm{B(:,s_i)}^2}\right)^\top \left(\hat{B} + \frac{B(:,s_i) (B(:,s_i)^\top B)}{\norm{B(:,s_i)}^2}\right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d691d56d7580112fdd2d2db8eb4cd4aa_l3.png)
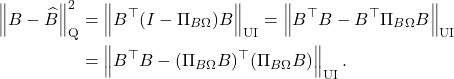
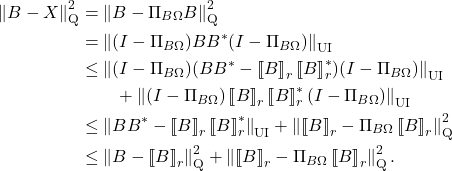
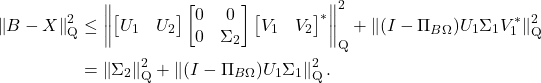
![Rendered by QuickLaTeX.com \begin{align*}\Pi_{B\Omega\Omega_1^\dagger} &= (B\Omega\Omega_1^\dagger) \left[(B\Omega\Omega_1^\dagger)^*(B\Omega\Omega_1^\dagger)\right]^\dagger (B\Omega\Omega_1^\dagger)^* \\&= (U_1\Sigma_1 + U_2\Sigma_2\Omega_2\Omega_1^\dagger) \left[\Sigma_1^2 + (\Sigma_2\Omega_2\Omega_1^\dagger)^*(\Sigma_2\Omega_2\Omega_1^\dagger)\right]^\dagger (U_1\Sigma_1 + U_2\Sigma_2\Omega_2\Omega_1^\dagger)^*.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d9e0cd585357787c460f6471f52ecb0a_l3.png)
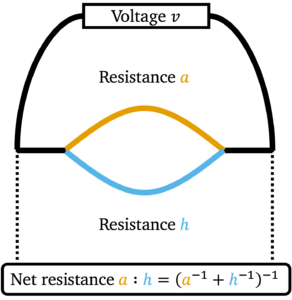
![Rendered by QuickLaTeX.com \[a:h \coloneqq \lim_{b\downarrow a, \: k\downarrow h} b:k = \begin{cases}\left( a^{-1}+h^{-1}\right)^{-1}, & a,h > 0 ,\\0, & \textrm{otherwise}.\end{cases}\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7c49cbcbcd8d8f75fa4f18a40562e718_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\left\|A:H\right\|_{\rm UI} &= \max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr((A:H)M) \\&= \max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr(M^{1/2}(A:H)M^{1/2})\\&= \max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr((M^{1/2}AM^{1/2}):(M^{1/2}HM^{1/2})) \\&\le \max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \left[\tr(M^{1/2}AM^{1/2}):\tr(M^{1/2}HM^{1/2}) \right] \\&\le \left[\max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr(M^{1/2}AM^{1/2})\right]:\left[\max_{M\succeq 0,\: \left\|M\right\|_{\rm UI}'\le 1} \tr(M^{1/2}HM^{1/2})\right]\\&= \left\|A\right\|_{\rm UI} : \left\|H\right\|_{\rm UI}.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4d5c2226a0ddf81d346da401ff4b53b9_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\left\|B - X\right\|_{\rm Q}^2&\le \left\| \Sigma_2 \right\|_{\rm Q}^2 + \left\|\Sigma_1^2 : [(\Sigma_2\Omega_2\Omega_1^\dagger)^*(\Sigma_2\Omega_2\Omega_1^\dagger)]\right\|_{\rm UI} \\&\le \left\| \Sigma_2 \right\|_{\rm Q}^2 + \left\|\Sigma_1^2\right\|_{\rm UI} : \left\|[(\Sigma_2\Omega_2\Omega_1^\dagger)^*(\Sigma_2\Omega_2\Omega_1^\dagger)]\right\|_{\rm UI} \\&= \left\| \Sigma_2 \right\|_{\rm Q}^2 + \left\|\Sigma_1\right\|_{\rm Q}^2 : \left\|\Sigma_2\Omega_2\Omega_1^\dagger\right\|^2_{\rm Q}.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-df07b0b9660af51129fd29552ad6c650_l3.png)
![Rendered by QuickLaTeX.com \[\mathbb{E} \left\|SGW\right\|_{\rm F}^2 = \mathbb{E} \sum_{ij} \left(\sum_{k\ell} s_{ik}g_{k\ell}w_{\ell j} \right)^2 = \mathbb{E} \sum_{ijk\ell} s_{ik}^2 w_{\ell j}^2 = \left\|S\right\|_{\rm F}^2\left\|W\right\|_{\rm F}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-dc219fd0dab2748965ecbf439c051db5_l3.png)
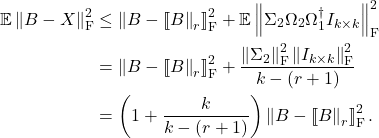
![Rendered by QuickLaTeX.com \[\mathbb{E} \left\|B - X\right\|^2 \le \min_{r \le k-2} \left( 1 + \frac{2r}{k-(r+1)} \right) \left(\sigma_{r+1}^2 + \frac{\mathrm{e}^2}{k-r} \sum_{i>r} \sigma_i^2 \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d4bb8b98ee0928e647c7cdbd91332fcc_l3.png)