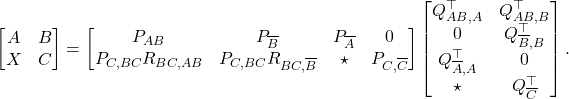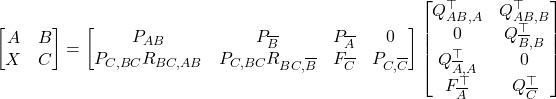I am delighted to share that me, Joel A. Tropp, and Robert J. Webber‘s paper XTrace: Making the Most of Every Sample in Stochastic Trace Estimation has recently been released as a preprint on arXiv. In it, we consider the implicit trace estimation problem:
Implicit trace estimation problem: Given access to a square matrix  via the matrix–vector product operation
via the matrix–vector product operation 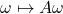 , estimate its trace
, estimate its trace 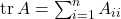 .
.
Algorithms for this task have many uses such as log-determinant computations in machine learning, partition function calculations in statistical physics, and generalized cross validation for smoothing splines. I described another application to counting triangles in a large network in a previous blog post.
Our paper presents new trace estimators XTrace and XNysTrace which are highly efficient, producing accurate trace approximations using a small budget of matrix–vector products. In addition, these algorithms are fast to run and are supported by theoretical results which explain their excellent performance. I really hope that you will check out the paper to learn more about these estimators!
For the rest of this post, I’m going to talk about the most basic stochastic trace estimation algorithm, the Girard–Hutchinson estimator. This seemingly simple algorithm exhibits a number of nuances and forms the backbone for more sophisticated trace estimates such as Hutch++, Nyström++, XTrace, and XNysTrace. Toward the end, this blog post will be fairly mathematical, but I hope that the beginning will be fairly accessible to all.
Girard–Hutchinson Estimator: The Basics
The Girard–Hutchinson estimator for the trace of a square matrix  is
is
(1) ![Rendered by QuickLaTeX.com \[\hat{\tr} = \frac{1}{m} \sum_{i=1}^m \omega_i^* A \omega_i. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-a287886ee24c9ab99dfe35b6cdae83cf_l3.png)
Here,  are random vectors, usually chosen to be statistically independent, and
are random vectors, usually chosen to be statistically independent, and  denotes the conjugate transpose of a vector or matrix. The Girard–Hutchinson estimator only depends on the matrix
denotes the conjugate transpose of a vector or matrix. The Girard–Hutchinson estimator only depends on the matrix  through the matrix–vector products
through the matrix–vector products  .
.
Unbiasedness
Provided the random vectors are isotropic
(2) ![Rendered by QuickLaTeX.com \[\mathbb{E} [\omega_i\omega_i^*] = I, \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-74bb76367ad1408bd538d049774cf990_l3.png)
the Girard–Hutchinson estimator is unbiased:
(3) ![Rendered by QuickLaTeX.com \[\mathbb{E} [\hat{\tr}] = \tr A.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f6407c7a741003cc20780e72d050cb56_l3.png)
Let us confirm this claim in some detail. First, we use linearity of expectation to evaluate
(4) ![Rendered by QuickLaTeX.com \[\mathbb{E} [\hat{\tr}] = \mathbb{E} \left[ \frac{1}{m} \sum_{i=1}^m \omega_i^*A\omega_i \right] = \frac{1}{m} \sum_{i=1}^m \mathbb{E} \left[ \omega_i^* A \omega_i\right]. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-52eebd003a597cac5b2c69f7b6e537e8_l3.png)
Therefore, to prove that ![Rendered by QuickLaTeX.com \mathbb{E} [\hat{\tr}] = \tr A](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-96f71a1b054ffed9e316cf9cb7a16c5c_l3.png) , it is sufficient to prove that
, it is sufficient to prove that ![Rendered by QuickLaTeX.com \mathbb{E} \left[\omega_i^*A\omega_i\right] = \tr A](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-120169dca277175c51f09b6f320cf5fa_l3.png) for each
for each 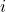 .
.
When working with traces, there are two tricks that solve 90% of derivations. The first trick is that, if we view a number as a 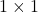 matrix, then a number equals its trace,
matrix, then a number equals its trace, 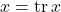 . The second trick is the cyclic property: For a
. The second trick is the cyclic property: For a 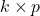 matrix
matrix 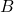 and a
and a 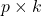 matrix
matrix 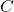 , we have
, we have 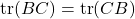 . The cyclic property should be handled with care when one works with a product of three or more matrices. For instance, we have
. The cyclic property should be handled with care when one works with a product of three or more matrices. For instance, we have
![Rendered by QuickLaTeX.com \[\tr[BCD] = \tr[(BC)D] = \tr[D(BC)] = \tr[DBC].\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-cc1cd4920a2431e93b5453b5373999e3_l3.png)
However,
![Rendered by QuickLaTeX.com \[\tr [BCD] \ne \tr[CBD] \quad \text{in general}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f563660fc58383a37ae7bedea16b2b9c_l3.png)
One should think of the matrix product 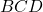 as beads on a closed loop of string. One can move the last bead
as beads on a closed loop of string. One can move the last bead 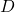 to the front of the other two,
to the front of the other two, ![Rendered by QuickLaTeX.com \tr [BCD] = \tr[DBC]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7656627b133add21ab8765532def07d6_l3.png) , but not interchange two beads,
, but not interchange two beads, ![Rendered by QuickLaTeX.com \tr[BCD] \ne \tr[CBD]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-0f43e4f72510a69b596f7b9f7504dd58_l3.png) .
.
With this trick in hand, let’s return to proving that ![Rendered by QuickLaTeX.com \mathbb{E} \left[\omega_i^*A\omega_i\right] = \tr A](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-120169dca277175c51f09b6f320cf5fa_l3.png) for every
for every 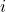 . Apply our two tricks:
. Apply our two tricks:
![Rendered by QuickLaTeX.com \[\mathbb{E} \left[\omega_i^*A\omega_i\right] = \mathbb{E} \tr \left[\omega_i^*A\omega_i\right] = \mathbb{E} \tr \left[A\omega_i\omega_i^*\right].\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-b1bb373766e64179bc0635f058a07f3f_l3.png)
The expectation is a linear operation and the matrix  is non-random, so we can bring the expectation into the trace as
is non-random, so we can bring the expectation into the trace as
![Rendered by QuickLaTeX.com \[\mathbb{E} \left[\omega_i^*A\omega_i\right] = \mathbb{E} \tr \left[A\omega_i\omega_i^*\right] = \tr(A \mathbb{E}[\omega_i\omega_i^*] ).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-1d10f835c99efab005f0ad12cdb5647b_l3.png)
Invoke the isotropy condition (2) and conclude:
![Rendered by QuickLaTeX.com \[\mathbb{E} \left[\omega_i^*A\omega_i\right] = \tr(A \mathbb{E}[\omega_i\omega_i^*] ) = \tr(A\cdot I) = \tr A.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-b51781498146df3547c1503e451ad682_l3.png)
Plugging this into (4) confirms the unbiasedness claim (3).
Variance
Continue to assume that the 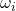 ‘s are isotropic (3) and now assume that
‘s are isotropic (3) and now assume that  are independent. By independence, the variance can be written as
are independent. By independence, the variance can be written as
![Rendered by QuickLaTeX.com \[\Var(\hat{\tr}) = \frac{1}{m^2} \sum_{i=1}^m \Var(\omega_i^*A\omega_i).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d9398dc43178ba9bb72aead0214e77c8_l3.png)
Assuming that  are identically distributed
are identically distributed 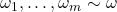 , we then get
, we then get
![Rendered by QuickLaTeX.com \[\Var(\hat{\tr}) = \frac{1}{m} \Var(\omega^*A\omega).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4501a18842234362f8f99d523c7c723e_l3.png)
The variance decreases like 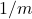 , which is characteristic of Monte Carlo-type algorithms. Since
, which is characteristic of Monte Carlo-type algorithms. Since 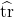 is unbiased (i.e, (3)), this means that the mean square error decays like
is unbiased (i.e, (3)), this means that the mean square error decays like 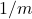 so the average error (more precisely root-mean-square error) decays like
so the average error (more precisely root-mean-square error) decays like
![Rendered by QuickLaTeX.com \[\left| \hat{\tr} - \tr A \right| \lessapprox \frac{\mathrm{const}}{\sqrt{m}}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-c735791b8fe88d627bc2ebad8137fb1e_l3.png)
This type of convergence is very slow. If I want to decrease the error by a factor of 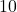 , I must do
, I must do 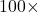 the work!
the work!
Variance-reduced trace estimators like Hutch++ and our new trace estimator XTrace improve the rate of convergence substantially. Even in the worst case, Hutch++ and XTrace reduce the variance at a rate 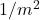 and (root-mean-square) error at rates
and (root-mean-square) error at rates 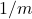 :
:
![Rendered by QuickLaTeX.com \[\Var(\hat{\tr}_{\text{H++ or X}}) \le \frac{\mathrm{const}}{m^2},\quad \left| \hat{\tr}_{\text{H++ or X}} - \tr A \right| \lessapprox \frac{\mathrm{const}}{m}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-87438d87094d3df389400b5f7abee24a_l3.png)
For matrices with rapidly decreasing singular values, the variance and error can decrease much faster than this.
Variance Formulas
As the rate of convergence for the Girard–Hutchinson estimator is so slow, it is imperative to pick a distribution on test vectors 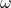 that makes the variance of the single–sample estimate
that makes the variance of the single–sample estimate 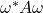 as low as possible. In this section, we will provide several explicit formulas for the variance of the Girard–Hutchinson estimator. Derivations of these formulas will appear at the end of this post. These variance formulas help illuminate the benefits and drawbacks of different test vector distributions.
as low as possible. In this section, we will provide several explicit formulas for the variance of the Girard–Hutchinson estimator. Derivations of these formulas will appear at the end of this post. These variance formulas help illuminate the benefits and drawbacks of different test vector distributions.
To express the formulas, we will need some notation. For a complex number 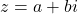 we use
we use 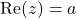 and
and 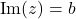 to denote the real and imaginary parts. The variance of a random complex number
to denote the real and imaginary parts. The variance of a random complex number  is
is
![Rendered by QuickLaTeX.com \[\Var(z) := \mathbb{E} |z - \mathbb{E} z|^2 = \Var(\Re z) + \Var(\Im z).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-8f636667a7dbbb3217896a962de1d28c_l3.png)
The Frobenius norm of a matrix  is
is
![Rendered by QuickLaTeX.com \[\left\|A\right\|_{\rm F}^2 = \sum_{i,j} |A_{ij}|^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-e132e4b9029c41dbfb99cd0e588d8d70_l3.png)
If  is real symmetric or complex Hermitian with (real) eigenvalues
is real symmetric or complex Hermitian with (real) eigenvalues  , we have
, we have
(5) ![Rendered by QuickLaTeX.com \[\left\|A\right\|_{\rm F}^2 = \sum_{i=1}^n \lambda_i^2. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-06b272e602552353c020f6399347c748_l3.png)
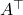 denotes the ordinary transpose of
denotes the ordinary transpose of  and
and 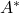 denotes the conjugate transpose of
denotes the conjugate transpose of  .
.
Real-Valued Test Vectors
We first focus on real-valued test vectors 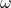 . Since
. Since 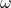 is real, we can use the ordinary transpose
is real, we can use the ordinary transpose  rather than the conjugate transpose
rather than the conjugate transpose  . Since
. Since 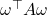 is a number, it is equal to its own transpose:
is a number, it is equal to its own transpose:
![Rendered by QuickLaTeX.com \[\omega^\top A \omega = (\omega^\top A \omega)^\top = \omega^\top A^\top \omega.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-3d543dc0d4d64d3c28f0f291cd9c8e83_l3.png)
Therefore,
![Rendered by QuickLaTeX.com \[\omega^\top A\omega = \frac{\omega^\top A \omega + \omega^\top A^\top \omega}{2} = \omega^\top \left( \frac{A + A^\top}{2} \right)\omega.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f1bbe4f0c983f73e2461b593b752f54a_l3.png)
The Girard–Hutchinson trace estimator applied to  is the same as the Girard–Hutchinson estimator applied to the symmetric part of
is the same as the Girard–Hutchinson estimator applied to the symmetric part of  ,
, 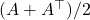 .
.
For the following results, assume  is symmetric,
is symmetric, 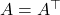 .
.
- Real Gaussian:
 are independent standard normal random vectors.
are independent standard normal random vectors. ![Rendered by QuickLaTeX.com \[\Var(\omega^\top A\omega) = 2 \left\|A\right\|_{\rm F}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-091cf638fab943be5fd224acdb42668f_l3.png)
- Uniform signs (Rademachers):
 are independent random vectors with uniform
are independent random vectors with uniform 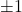 coordinates.
coordinates. ![Rendered by QuickLaTeX.com \[\Var(\omega^\top A \omega) = 2\sum_{i\ne j} |A_{ij}|^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-74621dad79cb3f06a8b70c661df4e01d_l3.png)
- Real sphere: Assume
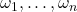 are uniformly distributed on the real sphere of radius
are uniformly distributed on the real sphere of radius 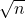 :
: 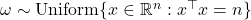 .
. ![Rendered by QuickLaTeX.com \[\Var(\omega^\top A\omega) = \frac{2n}{n+2} \left( \left\|A\right\|_{\rm F}^2 - \frac{1}{n} |\tr A|^2 \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-1e419b8da9b953a0b8b914df9e89f8c6_l3.png)
These formulas continue to hold for nonsymmetric  by replacing
by replacing  by its symmetric part
by its symmetric part 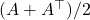 on the right-hand sides of these variance formulas.
on the right-hand sides of these variance formulas.
Complex-Valued Test Vectors
We now move our focus to complex-valued test vectors 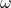 . As a rule of thumb, one should typically expect that the variance for complex-valued test vectors applied to a real symmetric matrix
. As a rule of thumb, one should typically expect that the variance for complex-valued test vectors applied to a real symmetric matrix  is about half the natural real counterpart—e.g., for complex Gaussians, you get about half the variance than with real Gaussians.
is about half the natural real counterpart—e.g., for complex Gaussians, you get about half the variance than with real Gaussians.
A square complex matrix has a Cartesian decomposition
![Rendered by QuickLaTeX.com \[A = A^{\rm H} + i A^{\rm SH}\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d61da43cd6497ceefd2306aabb52ee67_l3.png)
where
![Rendered by QuickLaTeX.com \[A^{\rm H} = \frac{A+A^*}{2} ,\quad A^{\rm SH} = \frac{A - A^*}{2i}\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-dc287a49a59dca9421fdd2bef6be5334_l3.png)
denote the Hermitian and skew-Hermitian parts of  . Similar to how the imaginary part of a complex number is real, the skew-Hermitian part of a complex matrix is Hermitian (and
. Similar to how the imaginary part of a complex number is real, the skew-Hermitian part of a complex matrix is Hermitian (and 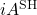 is skew-Hermitian). Since
is skew-Hermitian). Since 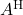 and
and 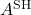 are both Hermitian, we have
are both Hermitian, we have
![Rendered by QuickLaTeX.com \[\Re(\omega^* A\omega) = \omega^* A^{\rm H} \omega, \quad \Im (\omega^* A \omega) = \omega^* A^{\rm SH} \omega.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-6809a35cd06d92218ce8d597847f74bd_l3.png)
Consequently, the variance of 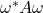 can be broken into Hermitian and skew-Hermitian parts:
can be broken into Hermitian and skew-Hermitian parts:
![Rendered by QuickLaTeX.com \[\Var(\omega^* A\omega) = \Var(\omega^* A^{\rm H}\omega) + \Var(\omega^* A^{\rm SH}\omega).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-e4658c38df479c5805dabd94b64cc0b3_l3.png)
For this reason, we will state the variance formulas only for Hermitian  , with the formula for general
, with the formula for general  following from the Cartesian decomposition.
following from the Cartesian decomposition.
For the following results, assume  is Hermitian,
is Hermitian, 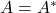 .
.
- Complex Gaussian:
 are independent standard complex random vectors, i.e., each
are independent standard complex random vectors, i.e., each 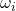 has iid entries distributed as
has iid entries distributed as 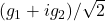 for
for 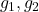 standard normal random variables.
standard normal random variables. ![Rendered by QuickLaTeX.com \[\Var(\omega^* A\omega) = \left\|A\right\|_{\rm F}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-1beb66e032d83431dac6f8e0d6655eec_l3.png)
- Uniform phases (Steinhauses):
 are independent random vectors whose entries are uniform on the complex unit circle
are independent random vectors whose entries are uniform on the complex unit circle 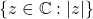 .
. ![Rendered by QuickLaTeX.com \[\Var(\omega^* A \omega) = \sum_{i\ne j} |A_{ij}|^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-1cb0e8df66c34056cd350752070af17a_l3.png)
- Complex sphere: Assume
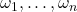 are uniformly distributed on the complex sphere of radius
are uniformly distributed on the complex sphere of radius 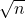 :
: 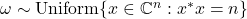 .
. ![Rendered by QuickLaTeX.com \[\Var(\omega^* A\omega) = \frac{n}{n+1} \left( \left\|A\right\|_{\rm F}^2 - \frac{1}{n} |\tr A|^2 \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-aacfa2afb45905f6abe0697c769cd1b7_l3.png)
Optimality Properties
Let us finally address the question of what the best choice of test vectors is for the Girard–Hutchinson estimator. We will state two results with different restrictions on  .
.
Our first result, due to Hutchinson, is valid for real symmetric matrices with real test vectors.
Optimality (independent test vectors with independent coordinates). If the test vectors 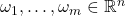 are isotropic (2), independent from each other, and have independent entries, then for any fixed real symmetric matrix
are isotropic (2), independent from each other, and have independent entries, then for any fixed real symmetric matrix  , the minimum variance for
, the minimum variance for 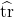 is obtained when
is obtained when  are populated with random signs
are populated with random signs 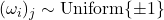 .
.
The next optimality results will have real and complex versions. To present the results for 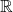 -valued and an
-valued and an 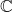 -valued test vectors on unified footing, let
-valued test vectors on unified footing, let 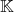 denote either
denote either 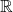 or
or 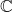 . We let a
. We let a 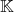 -Hermitian matrix be either a real symmetric matrix (if
-Hermitian matrix be either a real symmetric matrix (if 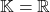 ) or a complex Hermitian matrix (if
) or a complex Hermitian matrix (if 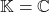 ). Let a
). Let a 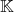 -unitary matrix be either a real orthogonal matrix (if
-unitary matrix be either a real orthogonal matrix (if 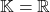 ) or a complex unitary matrix (if
) or a complex unitary matrix (if 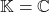 ).
).
The condition that the vectors  have independent entries is often too restrictive in practice. It rules out, for instance, the case of uniform vectors on the sphere. If we relax this condition, we get a different optimal distribution:
have independent entries is often too restrictive in practice. It rules out, for instance, the case of uniform vectors on the sphere. If we relax this condition, we get a different optimal distribution:
Optimality (independent test vectors). Consider any set 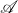 of
of 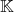 -Hermitian matrices which is invariant under
-Hermitian matrices which is invariant under 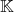 -unitary similary transformations:
-unitary similary transformations:
![Rendered by QuickLaTeX.com \[\text{If $A \in \mathscr{A}$ and $U$ is $\field$-unitary, then $U^*AU \in \mathscr{A}$.}\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-18b4216d44b2364ea39ed4a078a067af_l3.png)
Assume that the test vectors  are independent and isotropic (2). The worst-case variance
are independent and isotropic (2). The worst-case variance 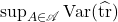 is minimized by choosing
is minimized by choosing  uniformly on the
uniformly on the 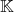 -sphere:
-sphere: 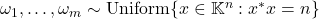 .
.
More simply, if you wants your stochastic trace estimator to be effective for a class of inputs 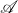 (closed under
(closed under 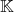 -unitary similarity transformations) rather than a single input matrix
-unitary similarity transformations) rather than a single input matrix  , then the best distribution are test vectors drawn uniformly from the sphere. Examples of classes of matrices
, then the best distribution are test vectors drawn uniformly from the sphere. Examples of classes of matrices 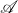 include:
include:
- Fixed eigenvalues. For fixed real eigenvalues
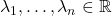 , the set of all
, the set of all 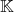 -Hermitian matrices with these eigenvalues.
-Hermitian matrices with these eigenvalues. - Density matrices. The class of all trace-one psd matrices.
- Frobenius norm ball. The class of all
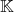 -Hermitian matrices of Frobenius norm at most 1.
-Hermitian matrices of Frobenius norm at most 1.
Derivation of Formulas
In this section, we provide derivations of the variance formulas. I have chosen to focus on derivations which are shorter but use more advanced techniques rather than derivations which are longer but use fewer tricks.
Real Gaussians
First assume  is real. Since
is real. Since  is real symmetric,
is real symmetric,  has an eigenvalue decomposition
has an eigenvalue decomposition 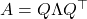 , where
, where 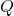 is orthogonal and
is orthogonal and 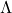 is a diagonal matrix reporting
is a diagonal matrix reporting  ‘s eigenvalues. Since the real Gaussian distribution is invariant under orthogonal transformations,
‘s eigenvalues. Since the real Gaussian distribution is invariant under orthogonal transformations, 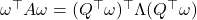 has the same distribution as
has the same distribution as 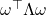 . Therefore,
. Therefore,
![Rendered by QuickLaTeX.com \[\Var(\omega^\top A \omega) = \Var(\omega^\top \Lambda \omega) = \Var \left( \sum_{i=1}^n \lambda_i \omega_i^2 \right) = \sum_{i=1}^n \lambda_i^2 \Var(\omega_i^2) = 2\sum_{i=1}^n \lambda_i^2 = 2\left\|A\right\|_{\rm F}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-a7416823720bac922fae7cc685892a49_l3.png)
Here, we used that the variance of a squared standard normal random variable is two.
For  non-real matrix, we can break the matrix
non-real matrix, we can break the matrix  into its entrywise real and imaginary parts
into its entrywise real and imaginary parts 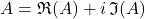 . Thus,
. Thus,
![Rendered by QuickLaTeX.com \[\Var(\omega^\top A \omega) = \Var(\omega^\top \mathfrak{R}(A) \omega) + \Var(\omega^\top \mathfrak{I}(A) \omega) = 2\left\|\mathfrak{R}(A)\right\|_{\rm F}^2 + 2\left\|\mathfrak{I}(A)\right\|_{\rm F}^2 = 2\left\|A\right\|_{\rm F}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-527980eb3510f2c015106e6945126d41_l3.png)
Uniform Signs
First, compute
![Rendered by QuickLaTeX.com \[\omega^\top A \omega - \mathbb{E}[\omega^\top A \omega] = \sum_{i,j=1}^n A_{ij} \omega_i\omega_j - \sum_{i=1}^n A_{ii} = \sum_{i\ne j} A_{ij} \omega_i\omega_j + \sum_{i=1}^n A_{ii}(\omega_i^2-1).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4489cc9cb97ee7a0896bc61193d79910_l3.png)
For a vector 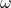 of uniform random signs, we have
of uniform random signs, we have 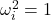 for every
for every 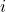 , so the second sum vanishes. Note that we have assumed
, so the second sum vanishes. Note that we have assumed  symmetric, so the sum over
symmetric, so the sum over 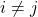 can be replaced by two times the sum over
can be replaced by two times the sum over 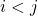 :
:
![Rendered by QuickLaTeX.com \[\omega^\top A \omega - \mathbb{E}[\omega^\top A \omega] = 2\sum_{i< j} A_{ij} \omega_i\omega_j.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-afc5f6bb40cd00b7897f25d73751f4ca_l3.png)
Note that 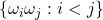 are pairwise independent. As a simple exercise, one can verify that the identity
are pairwise independent. As a simple exercise, one can verify that the identity
![Rendered by QuickLaTeX.com \[\Var(a_1 X_1+\cdots+a_kX_k) = |a_1|^2 \Var(X_1) + \cdots + |a_k|^2 \Var(X_k)\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-b6fd81d15f564f514f79ab4c33d133d8_l3.png)
holds for any pairwise independent family of random variances  and numbers
and numbers 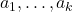 . Ergo,
. Ergo,
![Rendered by QuickLaTeX.com \begin{align*}\Var(\omega^\top A\omega) &= \Var(\omega^\top A \omega - \mathbb{E}[\omega^\top A \omega]) \\&= \Var\left(\sum_{i< j} 2A_{ij} \omega_i\omega_j\right) \\&= \sum_{i<j} 4 |A_{ij}|^2 \Var(\omega_i\omega_j) \\&= \sum_{i<j} 4 |A_{ij}|^2 \\&= 2 \sum_{i\ne j} |A_{ij}|^2.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-bbb33f378fabd391c28d938420d3a32b_l3.png)
In the second-to-last line, we use the fact that 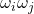 is a uniform random sign, which has variance
is a uniform random sign, which has variance 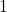 . The final line is a consequence of the symmetry of
. The final line is a consequence of the symmetry of  .
.
Uniform on the Real Sphere
The simplest proof is I know is by the “camel principle”. Here’s the story (a lightly edited quotation from MathOverflow):
A father left 17 camels to his three sons and, according to the will, the eldest son was to be given a half of the camels, the middle son one-third, and the youngest son the one-ninth. The sons did not know what to do since 17 is not evenly divisible into either two, three, or nine parts, but a wise man helped the sons: he added his own camel, the oldest son took 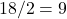 camels, the second son took
camels, the second son took 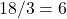 camels, the third son
camels, the third son 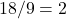 camels and the wise man took his own camel and went away.
camels and the wise man took his own camel and went away.
We are interested in a vector 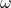 which is uniform on the sphere of radius
which is uniform on the sphere of radius 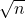 . Performing averages on the sphere is hard, so we add a camel to the problem by “upgrading”
. Performing averages on the sphere is hard, so we add a camel to the problem by “upgrading” 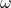 to a spherically symmetric vector
to a spherically symmetric vector 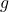 which has a random length. We want to pick a distribution for which the computation
which has a random length. We want to pick a distribution for which the computation 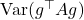 is easy. Fortunately, we already know such a distribution, the Gaussian distribution, for which we already calculated
is easy. Fortunately, we already know such a distribution, the Gaussian distribution, for which we already calculated 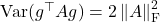 .
.
The Gaussian vector 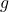 and the uniform vector
and the uniform vector 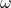 on the sphere are related by
on the sphere are related by
![Rendered by QuickLaTeX.com \[g = \sqrt{\frac{a}{n}} \omega,\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-a93345cf94749f98e715e42bc796501c_l3.png)
where  is the squared length of the Gaussian vector
is the squared length of the Gaussian vector 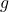 . In particular,
. In particular,  has the distribution of the sum of
has the distribution of the sum of 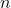 squared Gaussian random variables, which is known as a
squared Gaussian random variables, which is known as a 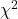 random variable with
random variable with 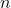 degrees of freedom.
degrees of freedom.
Now, we take the camel back. Compute the variance of 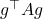 using the chain rule for variance:
using the chain rule for variance:
![Rendered by QuickLaTeX.com \[\Var(g^\top A g) = \mathbb{E}[\Var(g^\top A g \mid a)] + \Var(\mathbb{E}[g^\top A g \mid a]).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-e4e5ed235fbf5b2e026bcc58bd23becc_l3.png)
Here, 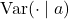 and
and ![Rendered by QuickLaTeX.com \mathbb{E}[ \cdot \mid a]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-6035d77442675ac2f790cfb5417e1072_l3.png) denote the conditional variance and conditional expectation with respect to the random variable
denote the conditional variance and conditional expectation with respect to the random variable  . The quick and dirty ways of working with these are to treat the random variable
. The quick and dirty ways of working with these are to treat the random variable  “like a constant” with respect to the conditional variance and expectation.
“like a constant” with respect to the conditional variance and expectation.
Plugging in the formula 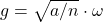 and treating
and treating  “like a constant”, we obtain
“like a constant”, we obtain
![Rendered by QuickLaTeX.com \begin{align*}\Var(g^\top A g) &= \mathbb{E}[\Var(a/n \cdot \omega^\top A \omega \mid a)] + \Var(\mathbb{E}[a/n \cdot \omega^\top A \omega \mid a]) \\&=\mathbb{E}[(a/n)^2\Var(\omega^\top A \omega)] + \Var(a/n \cdot \mathbb{E}[\omega^\top A \omega]) \\&= \frac{1}{n^2} \mathbb{E}[a^2] \cdot \Var(\omega^\top A \omega) + \frac{1}{n^2} \Var(a) |\mathbb{E} [\omega^\top A \omega]|^2.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-be052caddf1d08941925fa0283784635_l3.png)
As we mentioned,  is a
is a 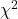 random variable with
random variable with 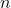 degrees of freedom and
degrees of freedom and ![Rendered by QuickLaTeX.com \mathbb{E}[a^2]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-53042ba72cdd3991b14b5d4508da6564_l3.png) and
and 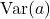 are known quantities that can be looked up:
are known quantities that can be looked up:
![Rendered by QuickLaTeX.com \[\mathbb{E}[a^2] = n(n+2), \quad \Var(a) = 2n.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-59ecda0ea21d92e43e3182d5cc5a7217_l3.png)
We know 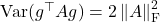 and
and ![Rendered by QuickLaTeX.com \mathbb{E} [\omega^\top A \omega] = \tr A](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-293908b9583df45d93349435d76b981a_l3.png) . Plugging these all in, we get
. Plugging these all in, we get
![Rendered by QuickLaTeX.com \[2\left\|A\right\|_{\rm F}^2 = \frac{n+2}{n} \Var(\omega^\top A\omega) + \frac{2}{n} |\tr A|^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-1c9b70fd4e5bc6ca4006114830d45619_l3.png)
Rearranging, we obtain
![Rendered by QuickLaTeX.com \[\Var(\omega^\top A\omega) = \frac{2n}{n+2} \left( \left\|A\right\|_{\rm F}^2 - \frac{1}{n}|\tr A|^2\right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7ab53e5895f9a1f474f5bf99fbec5662_l3.png)
Complex Gaussians
The trick is the same as for real Gaussians. By invariance of complex Gaussian random vectors under unitary transformations, we can reduce to the case where  is a diagonal matrix populated with eigenvalues
is a diagonal matrix populated with eigenvalues  . Then
. Then
![Rendered by QuickLaTeX.com \[\Var(\omega^*A \omega) = \Var \left( \sum_{i=1}^n \lambda_i |\omega_i|^2 \right) = \sum_{i=1}^n \Var(|\omega_i|^2) \lambda_i^2 = \sum_{i=1}^n \lambda_i^2 = \left\|A\right\|_{\rm F}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-68f4685a4cd845b98fc1a8fd1494e00a_l3.png)
Here, we use the fact that 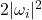 is a
is a 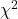 random variable with two degrees of freedom, which has variance four.
random variable with two degrees of freedom, which has variance four.
Random Phases
The trick is the same as for uniform signs. A short calculation (remembering that  is Hermitian and thus
is Hermitian and thus 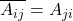 ) reveals that
) reveals that
![Rendered by QuickLaTeX.com \[\Var\left( \omega^* A \omega \right) = \Var \left( \sum_{i<j} 2 \Re(A_{ij} \overline{\omega_i} \omega_j) \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-b0e61723c22b056d1aca51694c407e6d_l3.png)
The random variables 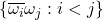 are pairwise independent so we have
are pairwise independent so we have
![Rendered by QuickLaTeX.com \[\Var\left( \omega^* A \omega \right) = \Var \left( \sum_{i<j} 2 \Re(A_{ij} \overline{\omega_i} \omega_j) \right) = 4\sum_{i<j} \Var \left( \Re(A_{ij} \overline{\omega_i} \omega_j) \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f2d77eb554a1e1ffbe4370efd8c704fb_l3.png)
Since 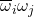 is uniformly distributed on the complex unit circle, we can assume without loss of generality that
is uniformly distributed on the complex unit circle, we can assume without loss of generality that 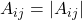 . Thus, letting
. Thus, letting 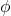 be uniform on the complex unit circle,
be uniform on the complex unit circle,
![Rendered by QuickLaTeX.com \[\Var\left( \omega^* A \omega \right) = 4\sum_{i<j} \Var \left( |A_{ij}|\Re(\phi)) \right) = 4\Var\left( \Re(\phi) \right)\sum_{i<j}|A_{ij}|^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-76d2ce84714c4549a86686259bd96847_l3.png)
The real and imaginary parts of 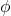 have the same distribution so
have the same distribution so
![Rendered by QuickLaTeX.com \[1 = \Var(\phi) = \Var(\Re \phi) + \Var(\Im \phi) = 2 \Var(\Re \phi)\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f83462ae4de80311482849de445edd5a_l3.png)
so 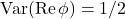 . Thus
. Thus
![Rendered by QuickLaTeX.com \[\Var\left( \omega^* A \omega \right) = 2 \sum_{i<j}|A_{ij}|^2 = \sum_{i\ne j} |A_{ij}|^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-436eeb79ad0eddde0752f1a398397ad5_l3.png)
Uniform on the Complex Sphere: Derivation 1 by Reduction to Real Case
There are at least three simple ways of deriving this result: the camel trick, reduction to the real case, and Haar integration. Each of these techniques illustrates a trick that is useful in its own right beyond the context of trace estimation. Since we have already seen an example of the camel trick for the real sphere, I will present the other two derivations.
Let us begin with the reduction to the real case. Let 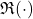 and
and 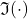 denote the real and imaginary parts of a vector or matrix, taken entrywise. The key insight is that if
denote the real and imaginary parts of a vector or matrix, taken entrywise. The key insight is that if 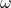 is a uniform random vector on the complex sphere of radius
is a uniform random vector on the complex sphere of radius 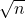 , then
, then
![Rendered by QuickLaTeX.com \[\mathscr{R}(\omega) := \twobyone{\mathfrak{R}(\omega)}{\mathfrak{I}(\omega)}\in\real^{2n} \quad \text{is a uniform random vector on the real sphere of radius $\sqrt{n}$}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-0ff873eff96e863cf02d36e65a5ca1be_l3.png)
We’ve converted the complex vector 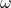 into a real vector
into a real vector 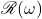 .
.
Now, we need to convert the complex matrix  into a real matrix
into a real matrix 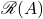 . To do this, recall that one way of representing complex numbers is by
. To do this, recall that one way of representing complex numbers is by 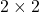 matrices:
matrices:
![Rendered by QuickLaTeX.com \[a + bi \iff \twobytwo{a}{-b}{b}{a}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-21b802836b33431568e5d854455d87d1_l3.png)
Using this correspondence addition and multiplication of complex numbers can be carried by addition and multiplication of the corresponding matrices.
To convert complex matrices to real matrices, we use a matrix-version of the same representation:
![Rendered by QuickLaTeX.com \[\mathscr{R}(A) = \twobytwo{\mathfrak{R}(A)}{-\mathfrak{I}(A)}{\mathfrak{I}(A)}{\mathfrak{R}(A)}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-12a37c4298c9e2d49dfa2f3ac32fe573_l3.png)
One can check that addition and multiplication of complex matrices can be carried out by addition and multiplication of the corresponding “realified” matrices, i.e.,
![Rendered by QuickLaTeX.com \[\mathscr{R}(A + B) = \mathscr{R}(A) + \mathscr{R}(B), \quad \mathscr{R}(A\cdot B) = \mathscr{R}(A) \cdot \mathscr{R}(B)\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4e7fe7da4edb58f1ee1bb8a2b1d2a78b_l3.png)
holds for all complex matrices  and
and 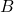 .
.
We’ve now converted complex matrix  and vector
and vector 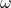 into real matrix
into real matrix 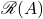 and vector
and vector 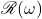 . Let’s compare
. Let’s compare 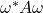 to
to 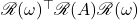 . A short calculation reveals
. A short calculation reveals
![Rendered by QuickLaTeX.com \[\omega^*A\omega = \mathscr{R}(\omega)^\top \mathscr{R}(A)\mathscr{R}(\omega) .\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d92927d567b8907621af83ab283672a5_l3.png)
Since 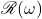 is a uniform random vector on the sphere of radius
is a uniform random vector on the sphere of radius 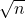 ,
, 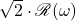 is a uniform random vector on the sphere of radius
is a uniform random vector on the sphere of radius 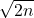 . Thus, by the variance formula for the real sphere, we get
. Thus, by the variance formula for the real sphere, we get
![Rendered by QuickLaTeX.com \[\Var(\omega^*A\omega) = \Var[(\sqrt{2}\mathscr{R}(\omega))^\top (\mathscr{R}(A)/2)(\sqrt{2}\mathscr{R}(\omega) )] = \frac{4n}{2n+2} \left[ \|\mathscr{R}(A)/2\|_{\rm F}^2 - \frac{1}{8n}(\tr\mathscr{R}(A))^2 \right].\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-ff16ba8dd015218ea642633d77f2a37a_l3.png)
A short calculation verifies that 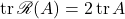 and
and 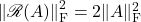 . Plugging this in, we obtain
. Plugging this in, we obtain
![Rendered by QuickLaTeX.com \[\Var(\omega^*A\omega)= \frac{n}{n+1} \left[ \|A\|_{\rm F}^2 - \frac{1}{n}(\tr A)^2 \right].\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-c9186eb813af3cd67173d73e32885a22_l3.png)
Uniform on the Complex Sphere: Derivation 2 by Haar Integration
The proof by reduction to the real case requires some cumbersome calculations and requires that we have already computed the variance in the real case by some other means. The method of Haar integration is more slick, but it requires some pretty high-power machinery. Haar integration may be a little bit overkill for this problem, but this technique is worth learning as it can handle some truly nasty expected value computations that appear, for example, in quantum information.
We seek to compute
![Rendered by QuickLaTeX.com \[\mathbb{E} [(\omega^*A \omega)^2].\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-8e3e10402f389b06dcd42db97c5ed116_l3.png)
The first trick will be to write this expession using a single matrix trace using the tensor (Kronecker) product 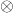 . For those unfamiliar with the tensor product, the main properties we will be using are
. For those unfamiliar with the tensor product, the main properties we will be using are
(6) ![Rendered by QuickLaTeX.com \[(A\otimes B) (C\otimes D) = (AB) \otimes (CD), \quad \tr(A\otimes B) = \tr A \cdot \tr B. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-c5ea1a0235766ad15b67d02b47cfea2e_l3.png)
We saw in the proof of unbiasedness that
![Rendered by QuickLaTeX.com \[\omega^* A \omega = \tr (\omega^*A\omega) = \tr (A \omega\omega^*).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-578bb521ecfb65fc4025f13431673b08_l3.png)
Therefore, by (6),
![Rendered by QuickLaTeX.com \[(\omega^*A\omega)^2 = (\tr [A \omega\omega^*])^2 = \tr [A\omega\omega^* \otimes A\omega\omega^*] = \tr [(A\otimes A) (\omega\omega^* \otimes \omega\omega^*)].\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-5161bc0d787be08130c4cfaed0c84489_l3.png)
Thus, to evaluate ![Rendered by QuickLaTeX.com \mathbb{E}[(\omega^*A\omega)^2]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-98fe9a5e8220c22709c9d6ade6c3141b_l3.png) , it will be sufficient to evaluate
, it will be sufficient to evaluate ![Rendered by QuickLaTeX.com \mathbb{E}[\omega\omega^* \otimes \omega\omega^*]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-06ac3d64041068176a86563cec4c67ba_l3.png) . Forunately, there is a useful formula for these expectation provided by a field of mathematics known as representation theory (see Lemma 1 in this paper):
. Forunately, there is a useful formula for these expectation provided by a field of mathematics known as representation theory (see Lemma 1 in this paper):
![Rendered by QuickLaTeX.com \[\mathbb{E}[ \omega\omega^* \otimes \omega\omega^*] = \frac{2n}{n+1} \operatorname{Proj}_{\operatorname{Sym}^2(\complex^n)}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-e5eddca3275c6bbb82fa6b789efe7664_l3.png)
Here, 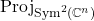 is the orthogonal projection onto the space of symmetric two-tensors
is the orthogonal projection onto the space of symmetric two-tensors 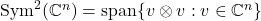 . Therefore, we have that
. Therefore, we have that
![Rendered by QuickLaTeX.com \[\mathbb{E}[(\omega^*A\omega)^2] = \tr [(A\otimes A) \mathbb{E}(\omega\omega^* \otimes \omega\omega^*)] = \frac{2n}{n+1} \tr [(A\otimes A) \operatorname{Proj}_{\operatorname{Sym}^2(\complex^n)}].\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-6c4f09e537808effd6c51c0aa4d20f6e_l3.png)
To evalute the trace on the right-hand side of this equation, there is another formula (see Lemma 6 in this paper):
![Rendered by QuickLaTeX.com \[\tr \left[(A\otimes B) \operatorname{Proj}_{\operatorname{Sym}^2(\complex^n)}\right] = \frac{1}{2} \left( \tr(AB) + \tr A \cdot \tr B \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-3ea7660d7e72964cf11ef2239731a497_l3.png)
Therefore, we conclude
![Rendered by QuickLaTeX.com \begin{align*}\Var(\omega^* A \omega) &= \mathbb{E}[(\omega^*A\omega)^2] - (\mathbb{E}[\omega^*A\omega])^2 \\&= \frac{2n}{n+1}\tr [(A\otimes A) \operatorname{Proj}_{\operatorname{Sym}^2(\complex^n)}] - (\tr A)^2 \\&= \frac{n}{n+1}\left[ \tr A^2 + (\tr A)^2 \right] - (\tr A)^2 \\&= \frac{n}{n+1}\left[ \left\|A\right\|_{\rm F}^2 - \frac{1}{n} (\tr A)^2 \right].\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7af313e9e07f20b93b0de83867f20a04_l3.png)
Proof of Optimality Properties
In this section, we provide proofs of the two optimality properties.
Optimality: Independent Vectors with Independent Coordinates
Assume  is real and symmetric and suppose that
is real and symmetric and suppose that 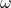 is isotropic (2) with independent coordinates. The isotropy condition
is isotropic (2) with independent coordinates. The isotropy condition
![Rendered by QuickLaTeX.com \[\mathbb{E}[\omega\omega^\top] = I\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-232c1e956304b14a7a6f4b55f9ba35e3_l3.png)
implies that ![Rendered by QuickLaTeX.com \mathbb{E}[\omega_i\omega_j] = \delta_{ij}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-9986528214c0653217367aca2a90d568_l3.png) , where
, where 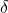 is the Kronecker symbol. Using this fact, we compute the second moment:
is the Kronecker symbol. Using this fact, we compute the second moment:
![Rendered by QuickLaTeX.com \begin{align*}\mathbb{E}[ (\omega^*A \omega)^2] &= \mathbb{E}\left[ \left( \sum_{i=1}^n A_{ii} \omega_i^2 +2 \sum_{i<j} A_{ij}\omega_i\omega_j) \right)^2\right] \\&= \sum_{i=1}^n A_{ii}^2 \mathbb{E}[\omega_i^4] + \sum_{i<j} (2A_{ii}A_{jj}+4A_{ij}^2) \mathbb{E}[\omega_i^2]\mathbb{E}[\omega_j^2] \\&= \sum_{i=1}^n A_{ii}^2 \mathbb{E}[\omega_i^4] + \sum_{i<j} (2A_{ii}A_{jj}+4A_{ij}^2) .\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4115914a6e648438e66028437f38ef2a_l3.png)
Thus
![Rendered by QuickLaTeX.com \[\Var(\omega^*A\omega) = \mathbb{E}[ (\omega^*A \omega)^2] - (\mathbb{E}[\omega^* A \omega])^2 = \sum_{i=1}^n A_{ii}^2 (\mathbb{E}[|\omega_i|^4]-1) + 4\sum_{i<j} A_{ij}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-006e2eb8d5f2162654d8c2de39f5a8bc_l3.png)
The variance is minimized by choosing 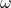 with
with 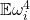 as small as possible. Since
as small as possible. Since 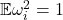 , the smallest possible value for
, the smallest possible value for 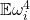 is
is 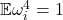 , which is obtained by populating
, which is obtained by populating 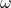 with random signs.
with random signs.
Optimality: Independent Vectors
This result appears to have first been proven by Richard Kueng in unpublished work. We use an argument suggested to me by Robert J. Webber.
Assume 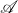 is a class of
is a class of 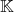 -Hermitian matrices closed under
-Hermitian matrices closed under 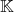 -unitary similarity transformations and that
-unitary similarity transformations and that 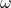 is an isotropic random vector (2). Decompose the test vector as
is an isotropic random vector (2). Decompose the test vector as
![Rendered by QuickLaTeX.com \[\omega = a \cdot s \quad \text{for} \quad a \in [0,+\infty), \: s \in\{x\in \field^n : x^*x = n \}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-11867b7bff40d97d79a74d5136774fd9_l3.png)
First, we shall show that the variance is reduced by replacing  with a vector
with a vector 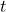 drawn uniformly from the sphere
drawn uniformly from the sphere
(7) ![Rendered by QuickLaTeX.com \[\sup_{A\in\mathscr{A}} \Var(\tilde{\omega}^*A\tilde{\omega}) \le \sup_{A\in\mathscr{A}} \Var(\omega^*A\omega \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-e54c170954072838232ab5ff7c78cf25_l3.png)
where
(8) ![Rendered by QuickLaTeX.com \[\tilde{\omega} = a\cdot t \quad \text{and}\quad t\sim \text{Uniform} \{ x \in \field^n :x^*x = n \} \quad \text{is independent of $a$}. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-cb668b2c9f168bca81deb51a17726b1d_l3.png)
Note that such a 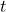 can be generated as
can be generated as 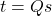 for a uniformly random
for a uniformly random 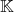 -unitary matrix
-unitary matrix 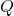 . Therefore, we have
. Therefore, we have
![Rendered by QuickLaTeX.com \begin{align*}\sup_{A\in\mathscr{A}} \Var(\tilde{\omega}^*A\tilde{\omega})&= \sup_{A\in\mathscr{A}} \left[\mathbb{E}[(\tilde{\omega}^*A\tilde{\omega})^2] - (\tr A)^2\right]\\&= \sup_{A\in\mathscr{A}} \left[\mathbb{E}[a^2 \cdot s^*(Q^*AQ)s] - (\tr (Q^*AQ))^2\right].\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-83564c7ba654655b42890c8adb65636f_l3.png)
Now apply Jensen’s inequality only over the randomness in 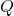 to obtain
to obtain
![Rendered by QuickLaTeX.com \begin{align*}\sup_{A\in\mathscr{A}} \Var(\tilde{\omega}^*A\tilde{\omega})&= \sup_{A\in\mathscr{A}} \left[\mathbb{E}[a^2 \cdot s^*(Q^*AQ)s] - (\tr (Q^*AQ))^2\right] \\&\le \mathbb{E}_Q \sup_{A\in\mathscr{A}} \left[\mathbb{E}_{a,s}[a^2 \cdot s^*(Q^*AQ)s] - (\tr (Q^*AQ))^2\right].\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f182f68bcb3cbe40c8b44cf5b585e474_l3.png)
Finally, note that since 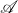 is closed under
is closed under 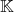 -unitary similarity transformations, the supremum over
-unitary similarity transformations, the supremum over 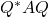 for
for 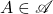 is the same as the supremum of
is the same as the supremum of 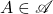 , so we obtain
, so we obtain
![Rendered by QuickLaTeX.com \begin{align*}\sup_{A\in\mathscr{A}} \Var(\tilde{\omega}^*A\tilde{\omega})&\le \mathbb{E}_Q \sup_{A\in\mathscr{A}} \left[\mathbb{E}_{a,s}[a^2 \cdot s^*(Q^*AQ)s] - (\tr (Q^*AQ))^2\right] \\&= \mathbb{E}_Q \sup_{A\in\mathscr{A}} \left[\mathbb{E}_{a,s}[a^2 \cdot s^*As] - (\tr A)^2\right] \\&= \sup_{A\in\mathscr{A}} \Var(\omega^*A\omega).\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-bd5736cb133d33ed28ab0f2305ac9678_l3.png)
We have successfully proven (7). This argument is a specialized version of a far more general result which appears as Proposition 4.1 in this paper.
Next, we shall prove
(9) ![Rendered by QuickLaTeX.com \[\sup_{A\in\mathscr{A}} \Var(t^*At) \le \sup_{A\in\mathscr{A}} \Var(\tilde{\omega}^*A\tilde{\omega}), \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-0de32ccbfe09388b9d36757b7f140a8e_l3.png)
where 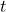 is still defined as in (8). Indeed, using the chain rule for variance, we obtain
is still defined as in (8). Indeed, using the chain rule for variance, we obtain
![Rendered by QuickLaTeX.com \begin{align*}\Var(\tilde{\omega}^*A\tilde{\omega})&= \Var(a^2\cdot t^*At) \\&= \mathbb{E}[\Var(a^2\cdot t^* A t \mid a)] + \Var(\mathbb{E}[a^2\cdot t^* A t \mid a]) \\&= \mathbb{E}[a^4]\Var(t^* A t )+ (\tr A)^2\Var(a^2) \\&\ge \mathbb{E}[a^4]\Var(t^* A t ).\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-c74465c627f5e70f943ce415fbfafd22_l3.png)
Here, we have used that 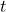 is uniform on the sphere and thus
is uniform on the sphere and thus ![Rendered by QuickLaTeX.com \mathbb{E}[t^*At] = \tr A](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-6ee9ad07ec8316cc6003116e3c311189_l3.png) . By definition,
. By definition,  is the length of
is the length of 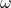 divided by
divided by 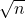 . Therefore,
. Therefore,
![Rendered by QuickLaTeX.com \[\mathbb{E}[a^2] = \frac{1}{n}\mathbb{E}[\omega^*\omega] = \frac{1}{n} \mathbb{E}[\tr (\omega\omega^*)] = \frac{1}{n} \tr (\mathbb{E}[\omega\omega^*]) = \frac{\tr I}{n} = 1.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4da8a9a33ffb6559da8625435549f7c2_l3.png)
Therefore, by Jensen’s inequality,
![Rendered by QuickLaTeX.com \[\mathbb{E}[a^4] = \mathbb{E}[(a^2)^2] \ge (\mathbb{E}[a^2])^2 = 1.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-356fd56d5d04dbda45fa992ae083be0f_l3.png)
Thus
![Rendered by QuickLaTeX.com \[\Var(\tilde{\omega}^*A\tilde{\omega}) \ge \mathbb{E}[a^4]\Var(t^* A t ) \ge \Var(t^*At) \quad \text{for every }A,\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-fc9331e3508e9102014ecc83f4517cc5_l3.png)
which proves (9).
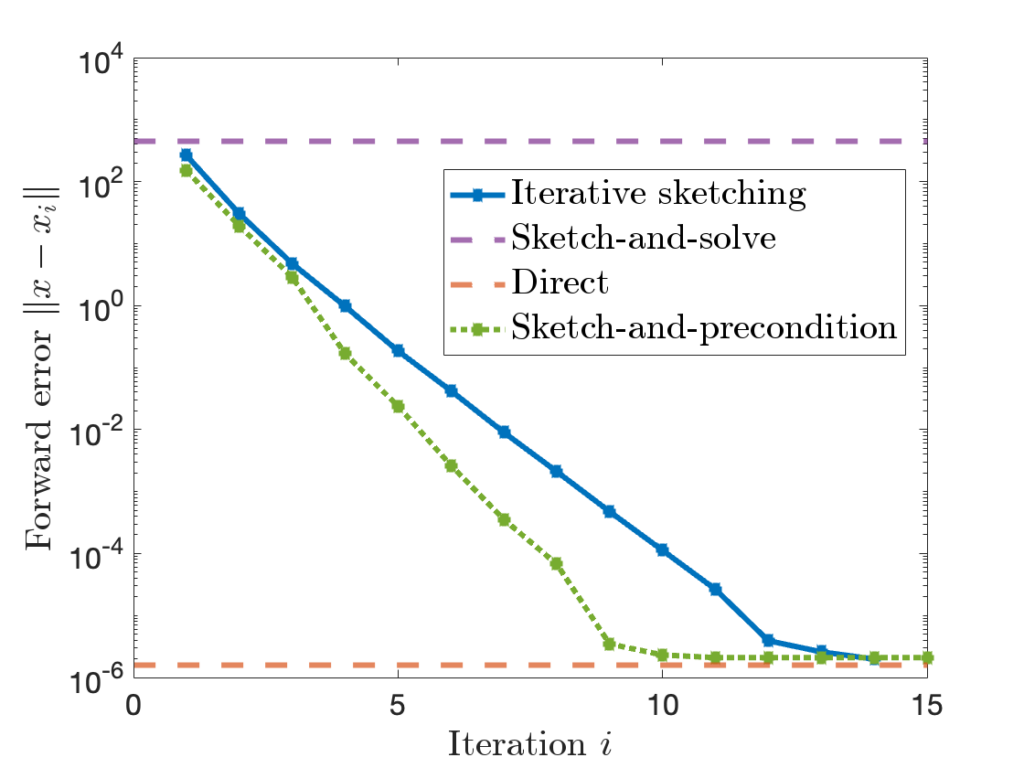
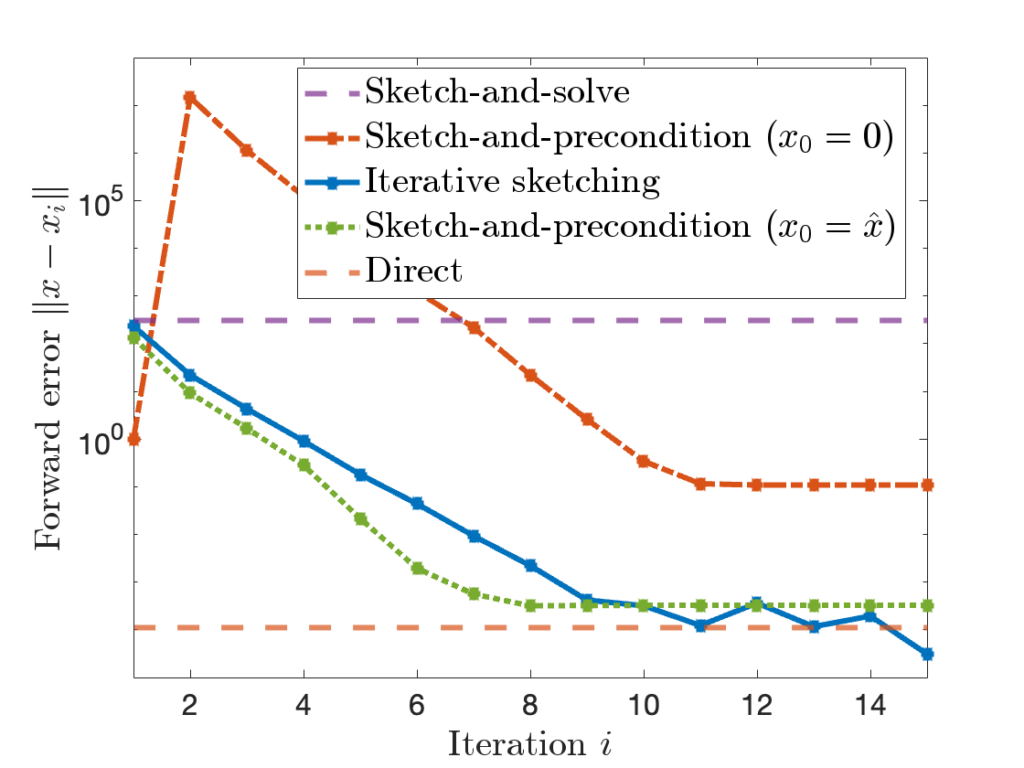
![]() approximations with a large block size of
approximations with a large block size of ![]() or a small block size of
or a small block size of ![]() , but these results give poor results for intermediate block sizes
, but these results give poor results for intermediate block sizes ![]() . But often these intermediate block sizes are the most efficient in practice. In our paper, we close this theoretical gap, showing RBKI is efficient for any block size
. But often these intermediate block sizes are the most efficient in practice. In our paper, we close this theoretical gap, showing RBKI is efficient for any block size ![]() . Check out the paper for details!
. Check out the paper for details!![]()
![]()
![]() be
be ![]() (distinct) input locations. If we evaluate
(distinct) input locations. If we evaluate ![]() at each number, we obtain a list of (output) values, which we denote by
at each number, we obtain a list of (output) values, which we denote by ![]() of
of ![]() (distinct) values, each of which given by the formula
(distinct) values, each of which given by the formula ![Rendered by QuickLaTeX.com \[p_a(\lambda_i) = \sum_{j=1}^t \lambda_i^{j-1} a_j.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-eaaf53acb1f30e1ced80af4704800e02_l3.png)
![]() . It is given by the formula
. It is given by the formula ![Rendered by QuickLaTeX.com \[V= \begin{bmatrix} 1 & \lambda_1 & \lambda_1^2 & \cdots & \lambda_1^{t-1} \\ 1 & \lambda_2 & \lambda_2^2 & \cdots & \lambda_2^{t-1} \\ 1 & \lambda_3 & \lambda_3^2 & \cdots & \lambda_3^{t-1} \\ \vdots & \vdots & \vdots & \ddots & \vdots \\ 1 & \lambda_s & \lambda_s^2 & \cdots & \lambda_s^{t-1} \end{bmatrix} \in \complex^{s\times t}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-2d100f803f43a4910ba6c03d2edde384_l3.png)
![]() so the number of locations
so the number of locations ![]() equals the number of coefficients
equals the number of coefficients ![]() . The Vandermonde matrix maps the vector of coefficients
. The Vandermonde matrix maps the vector of coefficients ![]() to the vector of values
to the vector of values ![]() . Its inverse
. Its inverse ![]() maps a set of values
maps a set of values ![]() to a set of coefficients
to a set of coefficients ![]() defining a polynomial
defining a polynomial ![]() which interpolates the values
which interpolates the values ![]() :
: ![]()
![]() , we first solve the linear system of equations
, we first solve the linear system of equations ![]() , obtaining a vector of coefficients
, obtaining a vector of coefficients ![]() . Then, define the interpolating polynomial
. Then, define the interpolating polynomial ![]()
![]() that is zero at the locations
that is zero at the locations ![]() but nonzero at the first location
but nonzero at the first location ![]() . (Remember that we have assumed that
. (Remember that we have assumed that ![]() are distinct.) Further, by rescaling this polynomial to
are distinct.) Further, by rescaling this polynomial to![]()
![]()
![Rendered by QuickLaTeX.com \[\ell_i(\lambda_j) = \delta_{ij} = \begin{cases} 1, & i = j, \\ 0, & i\ne j. \end{cases}\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-64c4effeb0f12225a76059d89ccce692_l3.png)
![]() , simply multiply each Lagrange polynomial
, simply multiply each Lagrange polynomial ![]() by the value
by the value ![]() and sum up, obtaining
and sum up, obtaining ![Rendered by QuickLaTeX.com \[q(u) = \sum_{i=1}^t f_i \ell_i(u).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-c1c5043718b5df795a883a6a60d26aaf_l3.png)
![Rendered by QuickLaTeX.com \[q(\lambda_j) = \sum_{i=1}^t f_i \ell_i(\lambda_j) = \sum_{i=1}^t f_i \delta_{ij} = f_j. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-90032b377ebe250ef36dccba2186b6ce_l3.png)
![]() as a linear combination of monomials
as a linear combination of monomials ![]() and the Lagrange formula (3) expresses
and the Lagrange formula (3) expresses ![]() as a linear combination of the Lagrange polynomials
as a linear combination of the Lagrange polynomials ![]() . To convert between these formulas, we just need to express the Lagrange polynomial basis in the monomial basis.
. To convert between these formulas, we just need to express the Lagrange polynomial basis in the monomial basis.![]() , and consider the fourth unnormalized Lagrange polynomial
, and consider the fourth unnormalized Lagrange polynomial ![]()
![]()
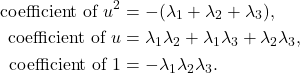
![]() . Specifically, given numbers
. Specifically, given numbers ![]() , the
, the ![]() th elementary symmetric polynomial
th elementary symmetric polynomial ![]() is defined as the sum of all products
is defined as the sum of all products ![]() of
of ![]() values, i.e.,
values, i.e., ![]()
![]() :
:![]() denote the list of locations without
denote the list of locations without ![]() . Then the
. Then the ![]() th Lagrange polynomial is given by
th Lagrange polynomial is given by ![Rendered by QuickLaTeX.com \[\ell_i(u) = \frac{\sum_{j=1}^t e_{t-j}(-\lambda_{-i})u^{j-1}}{\prod_{k\ne i} (\lambda_i - \lambda_k)}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-15d6cb602dcf916e14363b25a013870d_l3.png)
![Rendered by QuickLaTeX.com \[q(u) = \sum_{i=1}^t f_i\ell_i(u) = \sum_{i=1}^t \sum_{j=1}^t f_i \frac{e_{t-j}(-\lambda_{-i})}{\prod_{k\ne i} (\lambda_i - \lambda_k)} u^{j-1}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-17012971c8949a309b08ae230bf7fa94_l3.png)
![Rendered by QuickLaTeX.com \[q(u) = \sum_{j=1}^t \sum_{i=1}^t f_i \frac{e_{t-j}(-\lambda_{-i})}{\prod_{k\ne i} (\lambda_i - \lambda_k)} u^{j-1} = \sum_{j=1}^t \left(\sum_{i=1}^t \frac{e_{t-j}(-\lambda_{-i})}{\prod_{k\ne i} (\lambda_i - \lambda_k)}\cdot f_i \right) u^{j-1}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-10960bce48c6a124c253db4c1272f2e2_l3.png)
![Rendered by QuickLaTeX.com \[a_j = \sum_{i=1}^t \frac{e_{t-j}(-\lambda_{-i})}{\prod_{k\ne i} (\lambda_i - \lambda_k)}\cdot f_i.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-2bb3d1280c99ca124084f031ed343679_l3.png)
![]()
![]() can cause large changes to the coefficients
can cause large changes to the coefficients ![]() . On its face, this might seem like the problem of polynomial interpolation itself is ill-conditioned, but this is too hasty a conclusion. After all, it is only the mapping from values
. On its face, this might seem like the problem of polynomial interpolation itself is ill-conditioned, but this is too hasty a conclusion. After all, it is only the mapping from values ![]() to coefficients
to coefficients ![]() in the monomial basis that is ill-conditioned. Fortunately, there are much better, more numerically stable bases for representing a polynomial like the Chebyshev polynomials.
in the monomial basis that is ill-conditioned. Fortunately, there are much better, more numerically stable bases for representing a polynomial like the Chebyshev polynomials.![]() is. To characterize this ill-conditioning, we will utilize the condition number of the matrix
is. To characterize this ill-conditioning, we will utilize the condition number of the matrix ![]() . Given a norm
. Given a norm ![]() , the condition number of
, the condition number of ![]() is defined to be
is defined to be ![]()
![]()
![]()
![]() is straightforward. Indeed, setting
is straightforward. Indeed, setting ![]() and using (5), we compute
and using (5), we compute ![Rendered by QuickLaTeX.com \[\norm{V}_1= \max_{1\le j \le t} \sum_{i=1}^t |\lambda_i|^{j-1} \le \max_{1\le j \le t} tM^{j-1} = t\max\{1,M^{t-1}\}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-ec5f488cb8233df69dd22a00e9bae784_l3.png)
![]()
![]() . Fortunately, we have already done most of the hard work needed to bound this quantity. Using our expression (4) for the entries of
. Fortunately, we have already done most of the hard work needed to bound this quantity. Using our expression (4) for the entries of ![]() and using the formula (5) for the
and using the formula (5) for the ![]() -norm, we have
-norm, we have ![Rendered by QuickLaTeX.com \[\norm{\smash{V^{-1}}}_1= \max_{1\le j \le t} \sum_{i=1}^t |(V^{-1})_{ij}| = \max_{1\le j \le t}\frac{ \sum_{i=1}^t |e_{t-i}(-\lambda_{-j})|}{\prod_{k\ne j} |\lambda_j- \lambda_k|}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-b431915bf4dd9e0a3087cd91f5e3533a_l3.png)
![]()
![Rendered by QuickLaTeX.com \[\norm{\smash{V^{-1}}}_1\le \max_{1\le j \le t}\frac{ \sum_{i=1}^t e_{t-i}(|\lambda_{-j}|)}{\prod_{k\ne j} |\lambda_j- \lambda_k|}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-34c4b502a43c91513781e51c25190ad1_l3.png)
![]()
![]() as above, the numerator is bounded as
as above, the numerator is bounded as ![]() . To bound the denominator, let
. To bound the denominator, let ![]() be the smallest distance between two locations. Using
be the smallest distance between two locations. Using ![]() and
and ![]() , we can weaken the bound (6) to obtain
, we can weaken the bound (6) to obtain ![]()
![]()
and
. Then
![]() polynomial has precisely
polynomial has precisely ![]() roots. Consequently, at
roots. Consequently, at ![]() locations, it must be nonzero at least one. But how nonzero must the polynomial be at that one location? How large must it be? On this subject, the fundamental theorem of algebra is moot. However, Gautschi’s bound provides an answer.
locations, it must be nonzero at least one. But how nonzero must the polynomial be at that one location? How large must it be? On this subject, the fundamental theorem of algebra is moot. However, Gautschi’s bound provides an answer.![]() of the values
of the values ![]() ? Well, if we set all the coefficients
? Well, if we set all the coefficients ![]() to zero, then
to zero, then ![]() as well. So to avoid this trivial case, we should enforce a normalization condition on the coefficient vector
as well. So to avoid this trivial case, we should enforce a normalization condition on the coefficient vector ![]() , say
, say ![]() . With this setting, we are ready to compute. Begin by observing that
. With this setting, we are ready to compute. Begin by observing that ![]()
![]()
![]()
![]()
and its induced operator norm
, it holds that for any square invertible matrix
that
![]() on the values
on the values ![]() of a polynomial with coefficients
of a polynomial with coefficients ![]() . Combining with Gautschi’s bound gives the following robust version of the fundamental theorem of algebra:
. Combining with Gautschi’s bound gives the following robust version of the fundamental theorem of algebra:and locations
. Define
and
. Then
![]() locations, a degree-
locations, a degree-![]() polynomial must be nonzero at least one point. In fact, the sum of the values at these
polynomial must be nonzero at least one point. In fact, the sum of the values at these ![]() locations must be no worse than exponentially small in
locations must be no worse than exponentially small in ![]() .
.![Rendered by QuickLaTeX.com \[(u + \mu_1) (u + \mu_2) \cdots (u+\mu_k) = \sum_{j=0}^k e_{k-j}(\mu_1,\ldots,\mu_k) u^j.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-fe26dd7ba2e215fba4fea5cd28147cb2_l3.png)
![Rendered by QuickLaTeX.com \[\kappa_{\rm dem} = \frac{\norm{A}_{\rm F}}{\sigma_{\rm min}(A)} = \sqrt{\sum_i \left(\frac{\sigma_i(A)}{\sigma_{\rm min}(A)}\right)^2} \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-0a01bce740f28deced2d1a8b4e6e1002_l3.png)
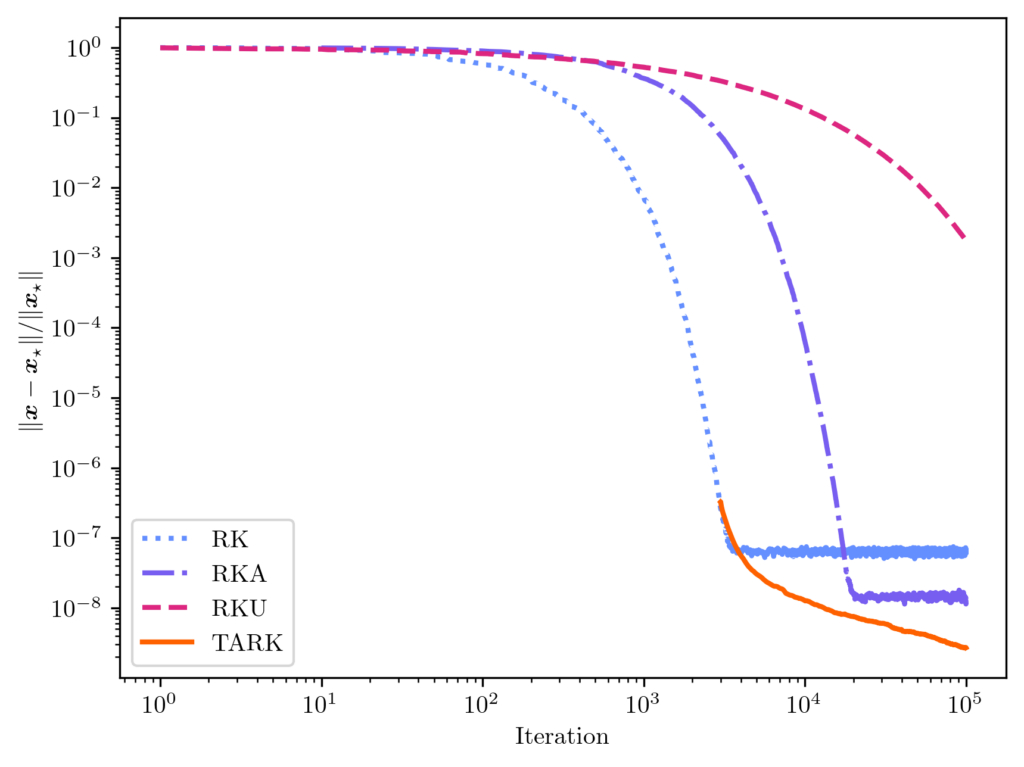
![Rendered by QuickLaTeX.com \[D \coloneqq \diag\left( \sqrt{\frac{p_j}{p_j^{\rm RK}}} : j =1,\ldots,n\right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-bc930770f7b4e20afffe903a4ab372ba_l3.png)
![Rendered by QuickLaTeX.com \[x_{t+1} = \left( I - \frac{a_{i_t}^{\vphantom{\top}}a_{i_t}^\top}{\norm{a_{i_t}}^2} \right)x_t + \frac{b_{i_t}a_{i_t}}{\norm{a_{i_t}}^2}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-cccfb54762bb990499c96331e7019309_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\expect_{i_t}[x_{t+1}] &= \sum_{j=1}^n \left[\left( I - \frac{a_j^{\vphantom{\top}} a_j^\top}{\norm{a_j}^2} \right)x_t + \frac{b_ja_j}{\norm{a_j}^2}\right] \prob\{i_t=j\}\\ &=\sum_{j=1}^n \left[\left( \frac{\norm{a_j}^2}{\norm{A}_{\rm F}^2}I - \frac{a_j^{\vphantom{\top}} a_j^\top}{\norm{A}_{\rm F}^2} \right)x_t + \frac{b_ja_j}{\norm{A}_{\rm F}^2}\right].\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-0a7f7b3bfad51b7094348db94d2d4254_l3.png)
![Rendered by QuickLaTeX.com \[\expect_{i_t}[x_{t+1}] = \left( I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right) x_t + \frac{A^\top b}{\norm{A}_{\rm F}^2}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-de4ded17daf001dde92c26025e9c14f3_l3.png)
![Rendered by QuickLaTeX.com \[\expect[x_{t+1}] = \left( I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right) \expect[x_t] + \frac{A^\top b}{\norm{A}_{\rm F}^2}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-24c5bfe6c1ea70293055deb68a3df117_l3.png)
![Rendered by QuickLaTeX.com \[\expect[x_t] = \left[\sum_{i=0}^{t-1} \left( I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^i \right]\cdot\frac{A^\top b}{\norm{A}_{\rm F}^2}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-eb47c96091b1187b53c318589b696693_l3.png)
![Rendered by QuickLaTeX.com \[\left(\frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^{-1} = \sum_{i=0}^\infty \left( I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^i. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-8d1be156303c168d2de83dbde76e44d6_l3.png)
![Rendered by QuickLaTeX.com \[\left(\frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^{-1} = \sum_{i=0}^{t-1} \left( I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^i + \sum_{i=t}^\infty \left( I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^i.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-08f4b1a705439315fe0fc038e059d01c_l3.png)
![Rendered by QuickLaTeX.com \[\expect[x_t] = x_\star - \left[\sum_{i=t}^\infty \left( I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^i \right]\cdot\frac{A^\top b}{\norm{A}_{\rm F}^2}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d973041dba49ba471bbfb6b3d4c2b463_l3.png)
![Rendered by QuickLaTeX.com \[\expect[x_t] - x_\star = - \left(I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^t \left[\sum_{i=0}^\infty \left( I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^i \right]\cdot\frac{A^\top b}{\norm{A}_{\rm F}^2}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-8238e65e7b7df2013504f2a6f71799e6_l3.png)
![Rendered by QuickLaTeX.com \[\expect[x_t] - x_\star = - \left(I - \frac{A^\top A}{\norm{A}_{\rm F}^2}\right)^t x_\star.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-3cbe72663f8ef13c762213b5592bb5d2_l3.png)
![Rendered by QuickLaTeX.com \[\norm{\expect[x_t] - x_\star}^2 \le \norm{I - \frac{A^\top A}{\norm{A}_{\rm F}^2}}^{2t} \norm{x}_\star^2. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-e663370119bd02900da0aa4f7b6faf03_l3.png)
![Rendered by QuickLaTeX.com \[I - \frac{A^\top A}{\norm{A}_{\rm F}^2} = I - V \cdot\frac{\Sigma^2}{\norm{A}_{\rm F}^2} \cdot V^\top = V \left( I - \frac{\Sigma^2}{\norm{A}_{\rm F}^2}\right)V^\top.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-468566feb9b39f9e0dd616b47f324ad5_l3.png)
![Rendered by QuickLaTeX.com \[\prob \{\text{any accepted this loop}\} = \sum_{i=1}^n \prob \{\text{$i$ accepted this loop}\} = \frac{\sum_{i=1}^n w_i}{\sum_{i=1}^n \rho_i}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-87004ae4b99269f6917de03d3a4ec86e_l3.png)
![Rendered by QuickLaTeX.com \[\sum_{j=1}^n k(s_i,s_j)w^\star_j = \int_\Omega k(s_i,x) g(x)\,\mathrm{d}\mu(x) \quad \text{for }i=1,2,\ldots,n.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-1a5ca36966f56dae9d994e208f9ef036_l3.png)
![Rendered by QuickLaTeX.com \[w\in\Delta\coloneqq \left\{ p\in\real^n_+ : \sum_{i=1}^n p_i = 1\right\}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-55c593b2593abc9ea8c18d4f8edbccd3_l3.png)
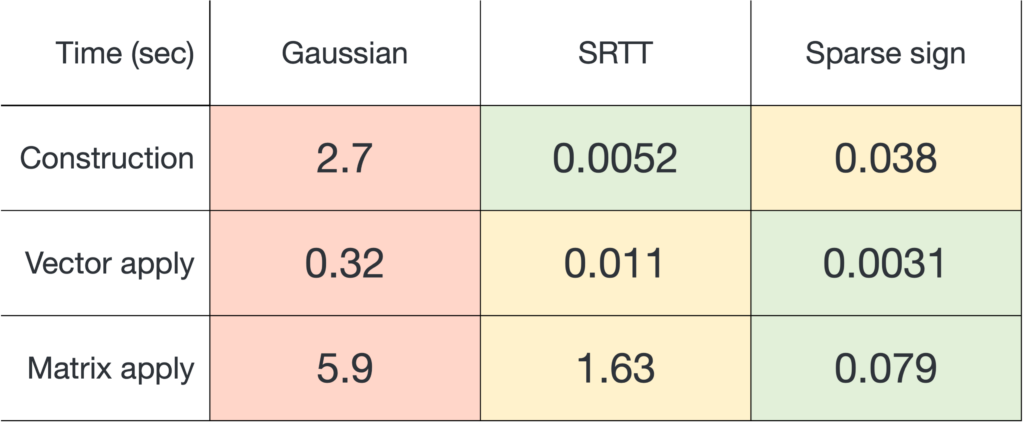
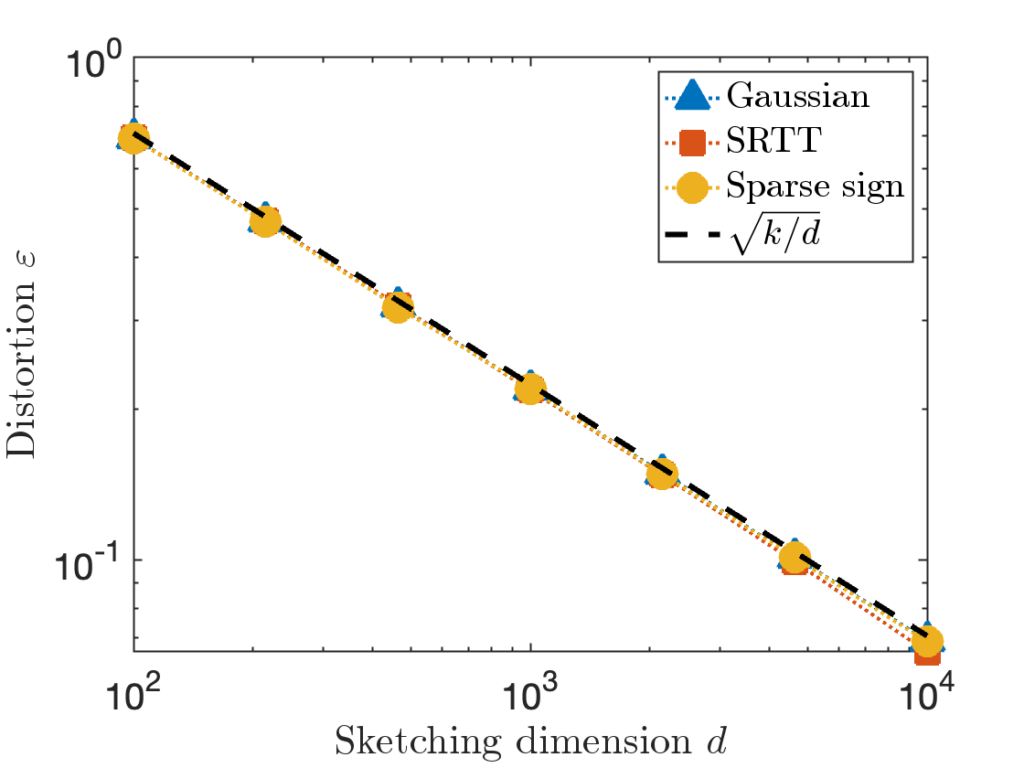
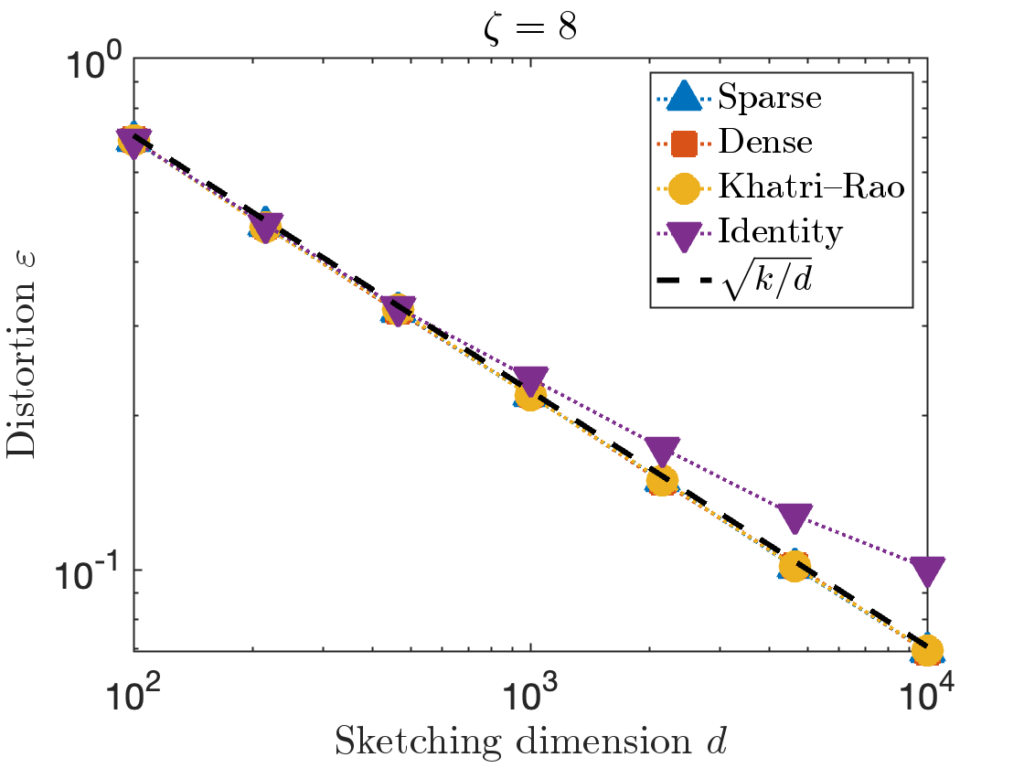
![Rendered by QuickLaTeX.com \[\zeta = \max \left( 8 , \left\lceil 2\sqrt{\frac{d}{k}} \right\rceil \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-5f1ebe7695d9918399b0f97b628fcce1_l3.png)
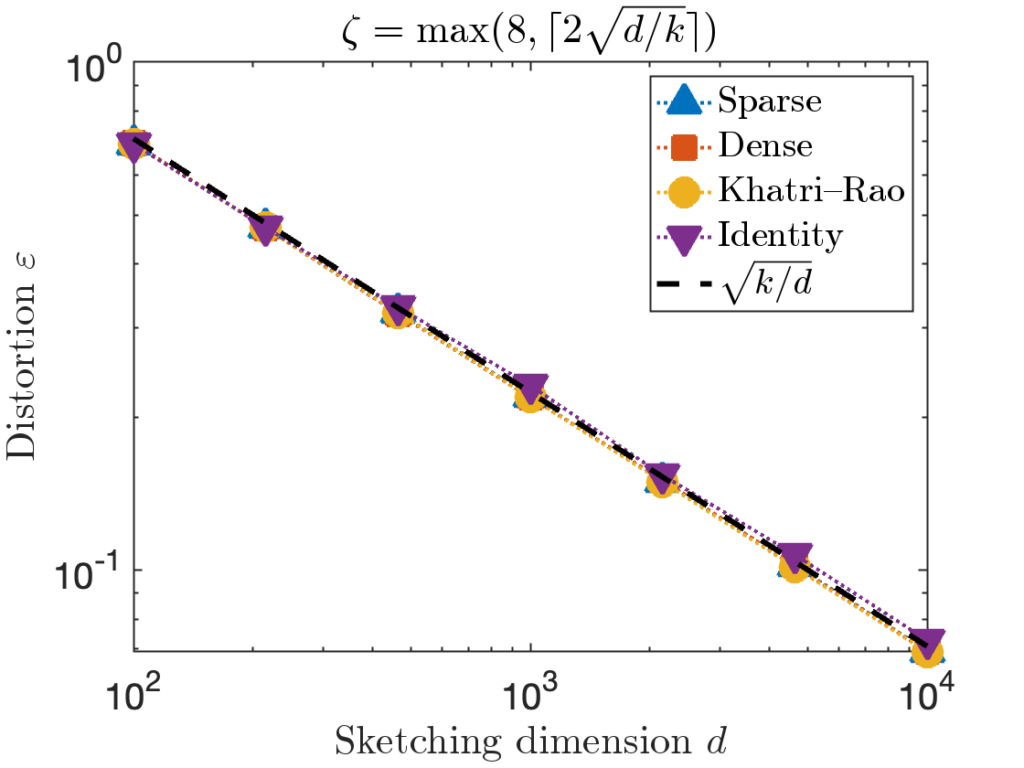
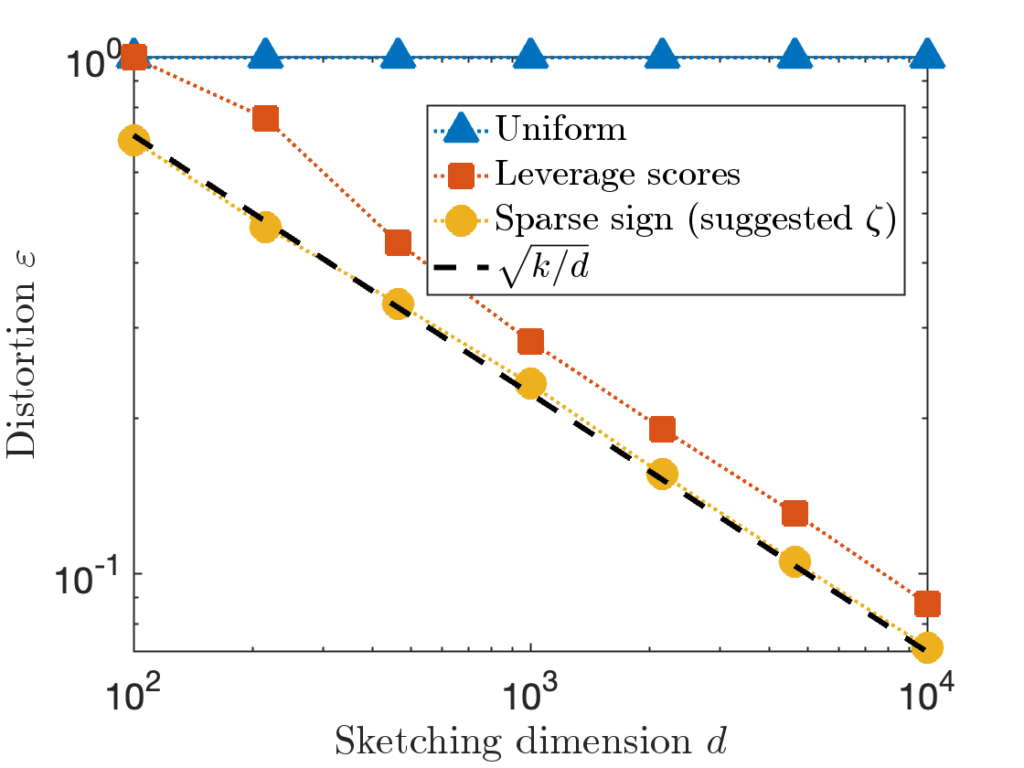
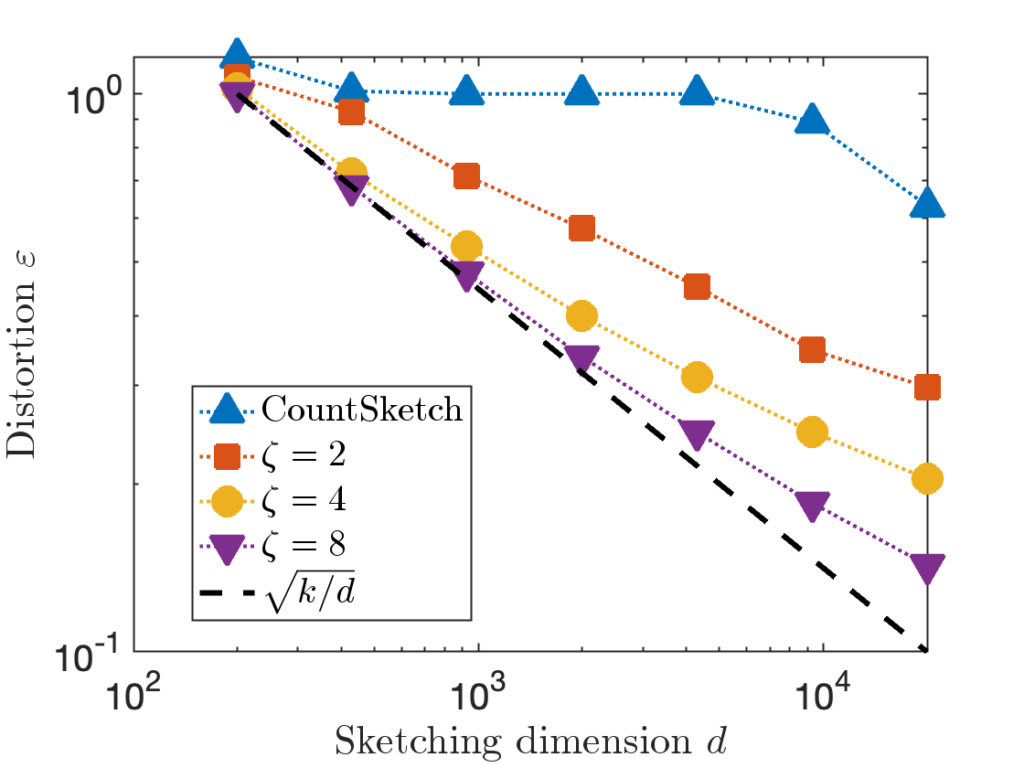
![Rendered by QuickLaTeX.com \[\prob \{ j_1,\ldots,j_k \text{ are distinct} \} = \prod_{i=1}^k \left(1 - \frac{i-1}{d} \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7dd28281f8dbed8b3a1452a21e7410c8_l3.png)
![Rendered by QuickLaTeX.com \[\mathbb{P} \{ j_1,\ldots,j_k \text{ are distinct} \} \le \prod_{i=0}^{k-1} \exp\left(-\frac{i}{d}\right) = \exp \left( -\frac{1}{d}\sum_{i=0}^{k-1} i \right) = \exp\left(-\frac{k(k-1)}{2d}\right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-8d9d4c7d12af0fd89e53ca261c5df287_l3.png)
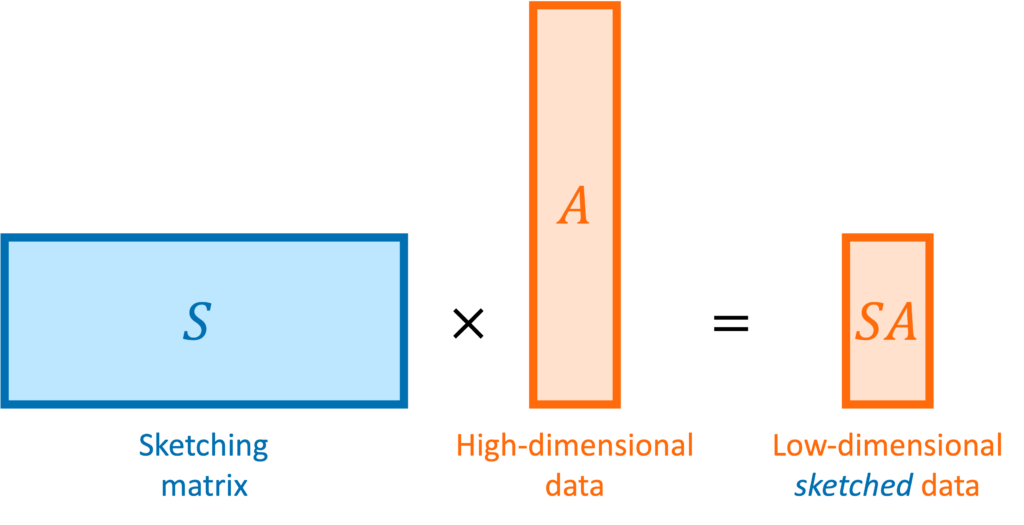
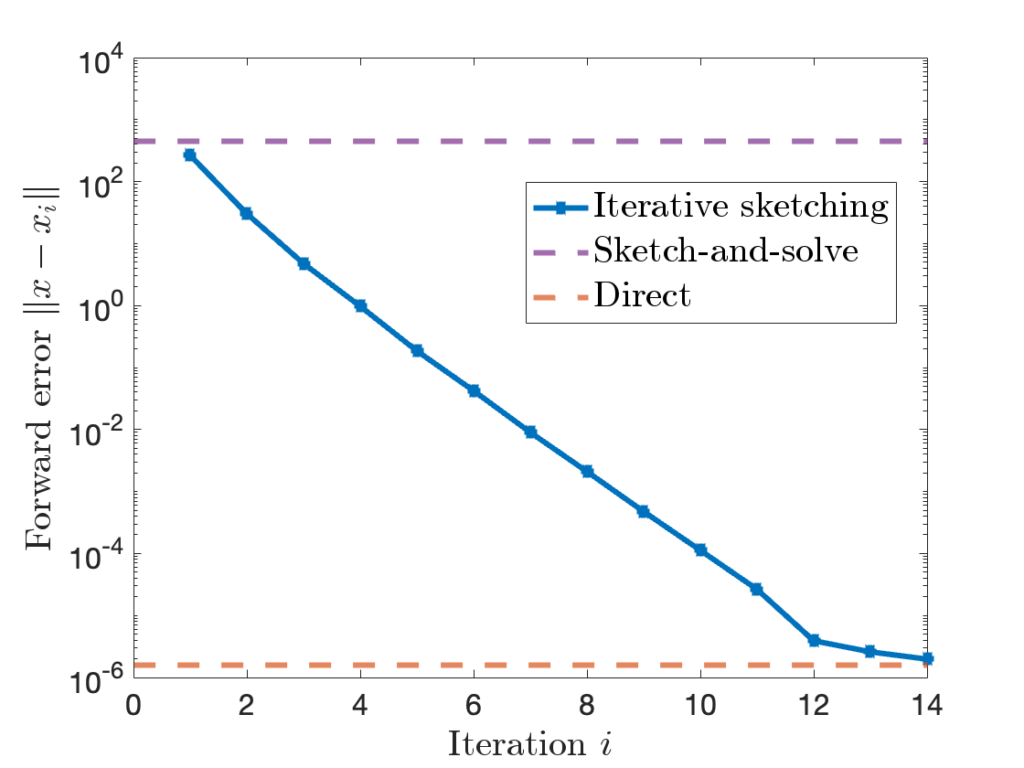
![Rendered by QuickLaTeX.com \[\mathbb{E} [\hat{\tr}] = \mathbb{E} \left[ \frac{1}{m} \sum_{i=1}^m \omega_i^*A\omega_i \right] = \frac{1}{m} \sum_{i=1}^m \mathbb{E} \left[ \omega_i^* A \omega_i\right]. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-52eebd003a597cac5b2c69f7b6e537e8_l3.png)
![Rendered by QuickLaTeX.com \[\Var(\omega^\top A \omega) = \Var(\omega^\top \Lambda \omega) = \Var \left( \sum_{i=1}^n \lambda_i \omega_i^2 \right) = \sum_{i=1}^n \lambda_i^2 \Var(\omega_i^2) = 2\sum_{i=1}^n \lambda_i^2 = 2\left\|A\right\|_{\rm F}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-a7416823720bac922fae7cc685892a49_l3.png)
![Rendered by QuickLaTeX.com \[\omega^\top A \omega - \mathbb{E}[\omega^\top A \omega] = \sum_{i,j=1}^n A_{ij} \omega_i\omega_j - \sum_{i=1}^n A_{ii} = \sum_{i\ne j} A_{ij} \omega_i\omega_j + \sum_{i=1}^n A_{ii}(\omega_i^2-1).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4489cc9cb97ee7a0896bc61193d79910_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\Var(\omega^\top A\omega) &= \Var(\omega^\top A \omega - \mathbb{E}[\omega^\top A \omega]) \\&= \Var\left(\sum_{i< j} 2A_{ij} \omega_i\omega_j\right) \\&= \sum_{i<j} 4 |A_{ij}|^2 \Var(\omega_i\omega_j) \\&= \sum_{i<j} 4 |A_{ij}|^2 \\&= 2 \sum_{i\ne j} |A_{ij}|^2.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-bbb33f378fabd391c28d938420d3a32b_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\Var(g^\top A g) &= \mathbb{E}[\Var(a/n \cdot \omega^\top A \omega \mid a)] + \Var(\mathbb{E}[a/n \cdot \omega^\top A \omega \mid a]) \\&=\mathbb{E}[(a/n)^2\Var(\omega^\top A \omega)] + \Var(a/n \cdot \mathbb{E}[\omega^\top A \omega]) \\&= \frac{1}{n^2} \mathbb{E}[a^2] \cdot \Var(\omega^\top A \omega) + \frac{1}{n^2} \Var(a) |\mathbb{E} [\omega^\top A \omega]|^2.\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-be052caddf1d08941925fa0283784635_l3.png)
![Rendered by QuickLaTeX.com \[\Var(\omega^*A \omega) = \Var \left( \sum_{i=1}^n \lambda_i |\omega_i|^2 \right) = \sum_{i=1}^n \Var(|\omega_i|^2) \lambda_i^2 = \sum_{i=1}^n \lambda_i^2 = \left\|A\right\|_{\rm F}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-68f4685a4cd845b98fc1a8fd1494e00a_l3.png)
![Rendered by QuickLaTeX.com \[\Var\left( \omega^* A \omega \right) = \Var \left( \sum_{i<j} 2 \Re(A_{ij} \overline{\omega_i} \omega_j) \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-b0e61723c22b056d1aca51694c407e6d_l3.png)
![Rendered by QuickLaTeX.com \[\Var\left( \omega^* A \omega \right) = \Var \left( \sum_{i<j} 2 \Re(A_{ij} \overline{\omega_i} \omega_j) \right) = 4\sum_{i<j} \Var \left( \Re(A_{ij} \overline{\omega_i} \omega_j) \right).\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f2d77eb554a1e1ffbe4370efd8c704fb_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\Var(\omega^* A \omega) &= \mathbb{E}[(\omega^*A\omega)^2] - (\mathbb{E}[\omega^*A\omega])^2 \\&= \frac{2n}{n+1}\tr [(A\otimes A) \operatorname{Proj}_{\operatorname{Sym}^2(\complex^n)}] - (\tr A)^2 \\&= \frac{n}{n+1}\left[ \tr A^2 + (\tr A)^2 \right] - (\tr A)^2 \\&= \frac{n}{n+1}\left[ \left\|A\right\|_{\rm F}^2 - \frac{1}{n} (\tr A)^2 \right].\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7af313e9e07f20b93b0de83867f20a04_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\mathbb{E}[ (\omega^*A \omega)^2] &= \mathbb{E}\left[ \left( \sum_{i=1}^n A_{ii} \omega_i^2 +2 \sum_{i<j} A_{ij}\omega_i\omega_j) \right)^2\right] \\&= \sum_{i=1}^n A_{ii}^2 \mathbb{E}[\omega_i^4] + \sum_{i<j} (2A_{ii}A_{jj}+4A_{ij}^2) \mathbb{E}[\omega_i^2]\mathbb{E}[\omega_j^2] \\&= \sum_{i=1}^n A_{ii}^2 \mathbb{E}[\omega_i^4] + \sum_{i<j} (2A_{ii}A_{jj}+4A_{ij}^2) .\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4115914a6e648438e66028437f38ef2a_l3.png)
![Rendered by QuickLaTeX.com \[\Var(\omega^*A\omega) = \mathbb{E}[ (\omega^*A \omega)^2] - (\mathbb{E}[\omega^* A \omega])^2 = \sum_{i=1}^n A_{ii}^2 (\mathbb{E}[|\omega_i|^4]-1) + 4\sum_{i<j} A_{ij}^2.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-006e2eb8d5f2162654d8c2de39f5a8bc_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\sup_{A\in\mathscr{A}} \Var(\tilde{\omega}^*A\tilde{\omega})&= \sup_{A\in\mathscr{A}} \left[\mathbb{E}[(\tilde{\omega}^*A\tilde{\omega})^2] - (\tr A)^2\right]\\&= \sup_{A\in\mathscr{A}} \left[\mathbb{E}[a^2 \cdot s^*(Q^*AQ)s] - (\tr (Q^*AQ))^2\right].\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-83564c7ba654655b42890c8adb65636f_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\sup_{A\in\mathscr{A}} \Var(\tilde{\omega}^*A\tilde{\omega})&= \sup_{A\in\mathscr{A}} \left[\mathbb{E}[a^2 \cdot s^*(Q^*AQ)s] - (\tr (Q^*AQ))^2\right] \\&\le \mathbb{E}_Q \sup_{A\in\mathscr{A}} \left[\mathbb{E}_{a,s}[a^2 \cdot s^*(Q^*AQ)s] - (\tr (Q^*AQ))^2\right].\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-f182f68bcb3cbe40c8b44cf5b585e474_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\sup_{A\in\mathscr{A}} \Var(\tilde{\omega}^*A\tilde{\omega})&\le \mathbb{E}_Q \sup_{A\in\mathscr{A}} \left[\mathbb{E}_{a,s}[a^2 \cdot s^*(Q^*AQ)s] - (\tr (Q^*AQ))^2\right] \\&= \mathbb{E}_Q \sup_{A\in\mathscr{A}} \left[\mathbb{E}_{a,s}[a^2 \cdot s^*As] - (\tr A)^2\right] \\&= \sup_{A\in\mathscr{A}} \Var(\omega^*A\omega).\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-bd5736cb133d33ed28ab0f2305ac9678_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\Var(\tilde{\omega}^*A\tilde{\omega})&= \Var(a^2\cdot t^*At) \\&= \mathbb{E}[\Var(a^2\cdot t^* A t \mid a)] + \Var(\mathbb{E}[a^2\cdot t^* A t \mid a]) \\&= \mathbb{E}[a^4]\Var(t^* A t )+ (\tr A)^2\Var(a^2) \\&\ge \mathbb{E}[a^4]\Var(t^* A t ).\end{align*}](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-c74465c627f5e70f943ce415fbfafd22_l3.png)
![Rendered by QuickLaTeX.com \[H = \begin{bmatrix} f_0 & f_1 & f_2 & \cdots & f_{(n-1)/2} \\f_1 & f_2 & f_3 & \cdots & f_{(n-1)/2+1} \\f_2 & f_3 & f_4 & \cdots & f_{(n-1)/2+2} \\\vdots & \vdots & \vdots & \ddots & \vdots \\f_{(n-1)/2} & f{(n-1)/2+1} & f{(n-1)/2+2} & \cdots & f_{n-1} \end{bmatrix}. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-e58f400c13a096efb5c1682c52003056_l3.png)
![Rendered by QuickLaTeX.com \[H_{ij} = f_{i+j} = \sum_{k=0}^5 d_k z_k^{i+j} = \sum_{k=0}^5 d_k z_k^i \cdot z_k^j. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-c66cda6c8df0e635353009a6469c9d13_l3.png)
![Rendered by QuickLaTeX.com \[V = \begin{bmatrix}z_0^0 & z_0^1 & z_0^2 & \cdots & z_0^{(n-1)/2} \\z_1^0 & z_1^1 & z_1^2 & \cdots & z_1^{(n-1)/2} \\\vdots & \vdots & \vdots & \ddots & \vdots \\z_5^0 & z_5^1 & z_5^2 & \cdots & z_5^{(n-1)/2}\end{bmatrix}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-52248d150045d64416cf49a569abf156_l3.png)
![Rendered by QuickLaTeX.com \[\begin{bmatrix} f_6 \\ f_7 \\ \vdots \\ f_{n-1} \end{bmatrix} = \underbrace{\begin{bmatrix} f_0 & f_1 & f_2 & f_3 & f_4 & f_5 \\ f_1 & f_2 & f_3 & f_4 & f_5 & f_6 \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ f_{n-7} & f_{n-6} & f_{n-5} & f_{n-4} & f_{n-3} & f_{n-2} \end{bmatrix}}_{=F}\begin{bmatrix} c_0 \\ c_1 \\ \vdots \\ c_5 \end{bmatrix}. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-7d7dc3e345c47a62fa233007d8d0db69_l3.png)
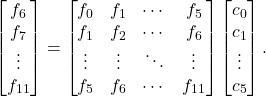 In particular, the matrix on the right-hand side of this equation is guaranteed to be nonsingular under our assumptions. Using the Vandermonde decomposition, can you see why?
In particular, the matrix on the right-hand side of this equation is guaranteed to be nonsingular under our assumptions. Using the Vandermonde decomposition, can you see why?![Rendered by QuickLaTeX.com \[\begin{bmatrix}f_0 \\ f_1 \\ \vdots \\ f_{n-1}\end{bmatrix} = \begin{bmatrix}1 & 1 & \cdots & 1 \\ z_0 & z_1 & \cdots & z_5 \\ \vdots & \vdots & \ddots & \vdots \\ z_0^{n-1} & z_1^{n-1} & \cdots & z_5^{n-1}\end{bmatrix} \begin{bmatrix}d_0 \\ d_1 \\ \vdots \\ d_{n-1}.\end{bmatrix}. \]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-4b18fe5a64350989e4534f4b08ef4c23_l3.png)
![Rendered by QuickLaTeX.com \[H^{(\ell)} = \begin{bmatrix} f_0^{(\ell)} & f_1^{(\ell)} & f_2^{(\ell)} & \cdots & f_{(n-1)/2}^{(\ell)} \\f_1^{(\ell)} & f_2^{(\ell)} & f_3^{(\ell)} & \cdots & f_{(n-1)/2+1}^{(\ell)} \\f_2^{(\ell)} & f_3^{(\ell)} & f_4^{(\ell)} & \cdots & f_{(n-1)/2+2}^{(\ell)} \\\vdots & \vdots & \vdots & \ddots & \vdots \\f_{(n-1)/2}^{(\ell)} & f_{(n-1)/2+1}^{(\ell)} & f_{(n-1)/2+2}^{(\ell)} & \cdots & f_{n-1}^{(\ell)} \end{bmatrix}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-426cddcdbfa1b1bae2c3aaccd49f9974_l3.png)
 This matrix has rank-two but no rank-two confluent Vandermonde decomposition. The issue is that when extended to an infinite Hankel matrix
This matrix has rank-two but no rank-two confluent Vandermonde decomposition. The issue is that when extended to an infinite Hankel matrix  this (infinite!) matrix has a rank exceeding the size of the original Hankel matrix
this (infinite!) matrix has a rank exceeding the size of the original Hankel matrix ![Rendered by QuickLaTeX.com \[T = \begin{bmatrix} f_{(n-1)/2} & f_{(n-1)/2+1} & f_{(n-1)/2+2} & \cdots & f_{n-1} \\\\f_{(n-1)/2-1} & f_{(n-1)/2} & f_{(n-1)/2+1} & \cdots & f_{n-2} \\\\f_{(n-1)/2-2} & f_{(n-1)/2-1} & f_{(n-1)/2} & \cdots & f_{n-3} \\\\\vdots & \vdots & \vdots & \ddots & \vdots \\\\ f_0 & f_1 & f_2 & \cdots & f_{(n-1)/2} \end{bmatrix}.\]](https://www.ethanepperly.com/wp-content/ql-cache/quicklatex.com-d576130eb33a8d98958a967ae78ae988_l3.png)